This is a continuation of my past articles on protein binder design. Here I'll cover the state-of-the-art in AI antibody design.
Antibodies and antibody fragments (e.g., Fab, scFv, VHH) are particularly important in biotech, because they are highly specific, adaptable to almost any target, and have a proven track record as therapeutics. Full antibodies also have Fc regions, so they can activate the immune system as well as bind. In this article I'll just use the term "antibody" but many of the design approaches discussed below generate these smaller antibody fragments.
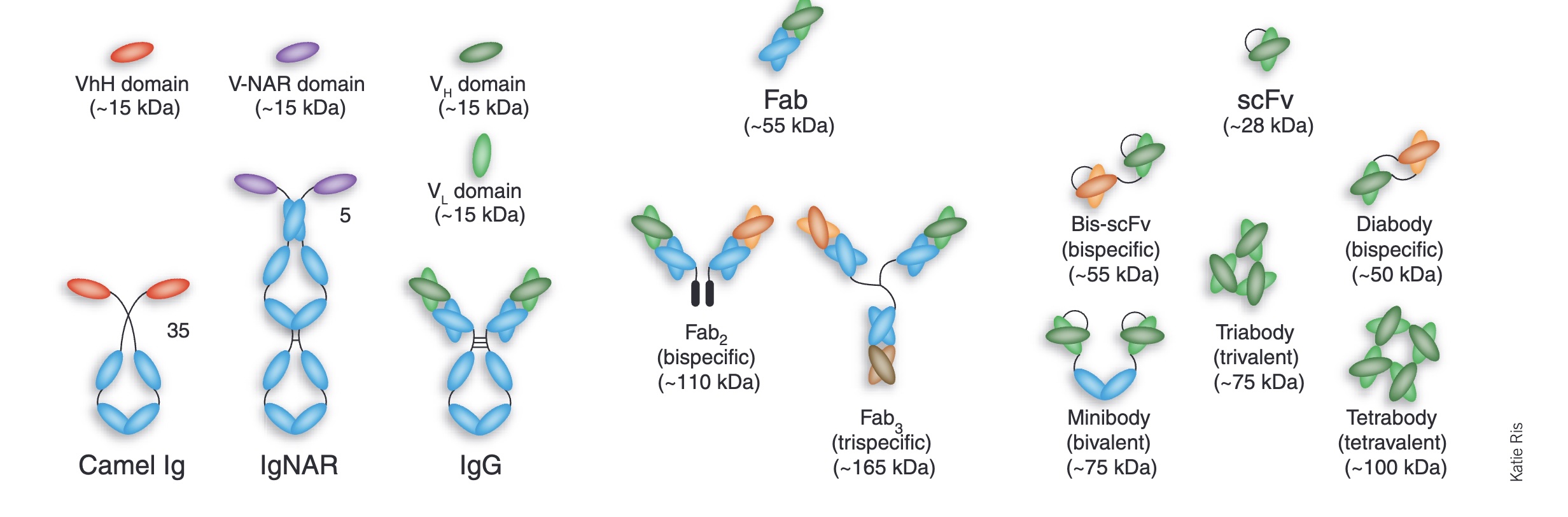
A menagerie of antibody fragments (Engineered antibody fragments and the rise of single domains, Nature Biotech, 2005)
Last year we saw a lot of progress in mini-binder design (especially BindCraft), but this year there has been a lot of activity in antibody and peptide design too, as it becomes clear that there are commercially important opportunities here. BindCraft 2 will likely include the ability to create antibody fragments; a fork called FoldCraft already enables this.
Antibodies are proteins, so why is antibody design not just the same problem as mini-binder design? In most ways they are the same. The main difference is that the CDR loops that drive antibody binding are highly variable and do not benefit directly from evolutionary information the way other binding motifs do. Folding long CDR loops correctly is especially difficult.
Here I'll review the latest antibody design tools. I'll also provide some biomodals code to run in case the reader wants to actually design their own antibodies!
RFantibody
While there were other antibody design tools before it, especially antibody language models, RFantibody was arguably the first successful de novo antibody design model. It is a fine-tuned variant of RFdiffusion, and like RFdiffusion it requires testing thousands of designs to have a good shot at producing a binder. The original RFantibody paper was originally published way back in March 2024, so as you'd expect, the performance—while remarkable for the time—has been surpassed, and Baker lab seems to have moved on to the next challenge. (Note, the preprint was first published in 2024 but the code was only released this year.)
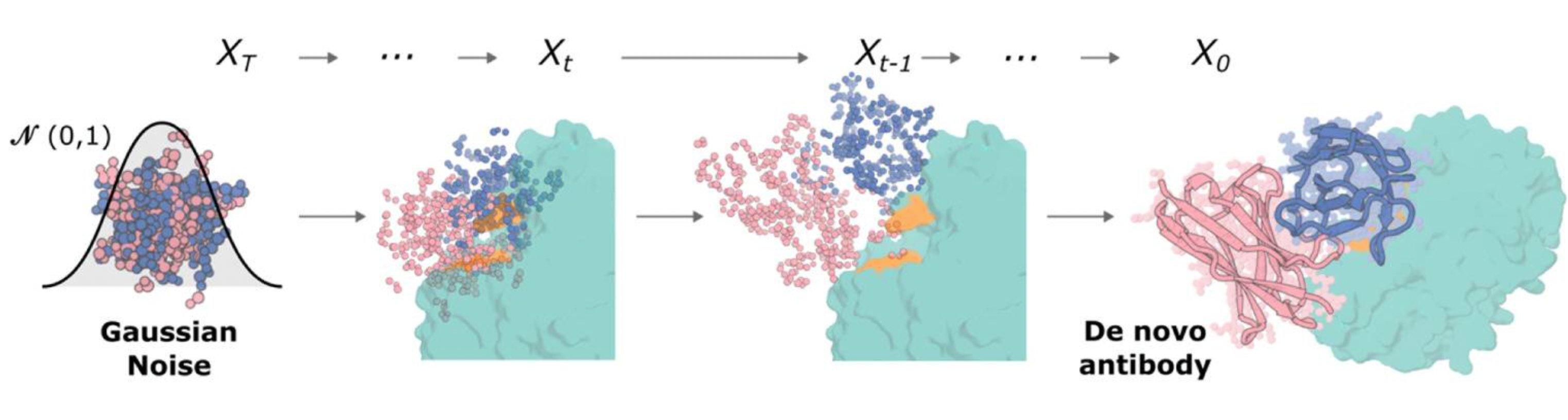
The diffusion process as illustrated in the RFantibody paper
IgGM
It's pretty interesting how many Chinese protein models there are now. Many of these models are from random internet companies just flexing their AI muscles.
IgGM is a brand new, comprehensive antibody design suite from Tencent (the giant internet conglomerate). It can do de novo design, affinity maturation, and more.
There are some troubling aspects to the IgGM paper. Diego del Alamo notes that the plots have unrealistically low variance (see the suspicious-looking plot below). When I run the code, I see what look like not-fully-folded structures. However, there is also strong empirical evidence it's a good model: a third place finish in the AIntibody competition (more information on that below).
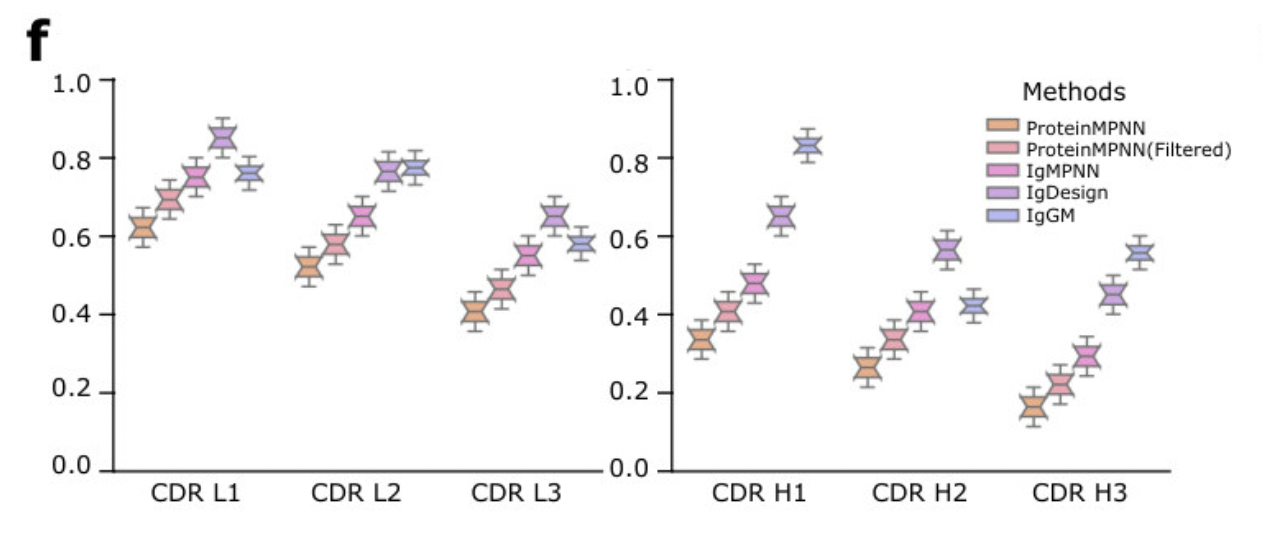
Suspiciously tight distributions in plots from the IgGM paper.
Sometimes this is due to plotting standard error vs standard deviation.
To run IgGM and generate a nanobody for PD-L1, run the following code:
# get the PD-L1 model from the Chai-2 technical report, only the A chain
curl -s https://files.rcsb.org/download/5O45.pdb | grep "^ATOM.\{17\}A" > 5O45_chainA.pdb
# get a nanobody sequence from 3EAK; replace CDR3 with Xs; tack on the sequence of 5O45 chain A
echo ">H\nQVQLVESGGGLVQPGGSLRLSCAASGGSEYSYSTFSLGWFRQAPGQGLEAVAAIASMGGLTYYADSVKGRFTISRDNSKNTLYLQMNSLRAEDTAVYYCXXXXXXXXXWGQGTLVTVSSRGRHHHHHH\n>A\nNAFTVTVPKDLYVVEYGSNMTIECKFPVEKQLDLAALIVYWEMEDKNIIQFVHGEEDLKVQHSSYRQRARLLKDQLSLGNAALQITDVKLQDAGVYRCMISYGGADYKRITVKVNAPYAAALEHHHHHH" > binder_X.fasta
# run IgGM; use the same hotspot from the Chai-2 technical report (add --relax for pyrosetta relaxation)
uvx modal run modal_iggm.py --input-fasta binder_X.fasta --antigen 5O45_chainA.pdb --epitope 56,115,123 --task design --run-name 5O45_r1
IgGM has one closed library dependency, PyRosetta, but this is only used for relaxing the final design, so it is optional. There are other ways to relax the structure, like using pr_alternative_utils.py from FreeBindCraft (a fork of BindCraft that does not depend on PyRosetta) or openmm via biomodals as shown below. FreeBindCraft's relax step has extra safeguards that likely make it work better than the code below.
uvx modal run modal_md_protein_ligand.py --pdb-id out/iggm/5O45_r1/input_0.pdb --num-steps 50000
PXDesign
Speaking of Chinese models, there is also a new mini-binder design tool called PXDesign from ByteDance, which is available for commercial use, but only via a server. It came out of beta just this week. The claimed performance is excellent, comparable to Chai-2. (The related Protenix protein structure model, "a trainable, open-source PyTorch reproduction of AlphaFold 3", is fully open.)
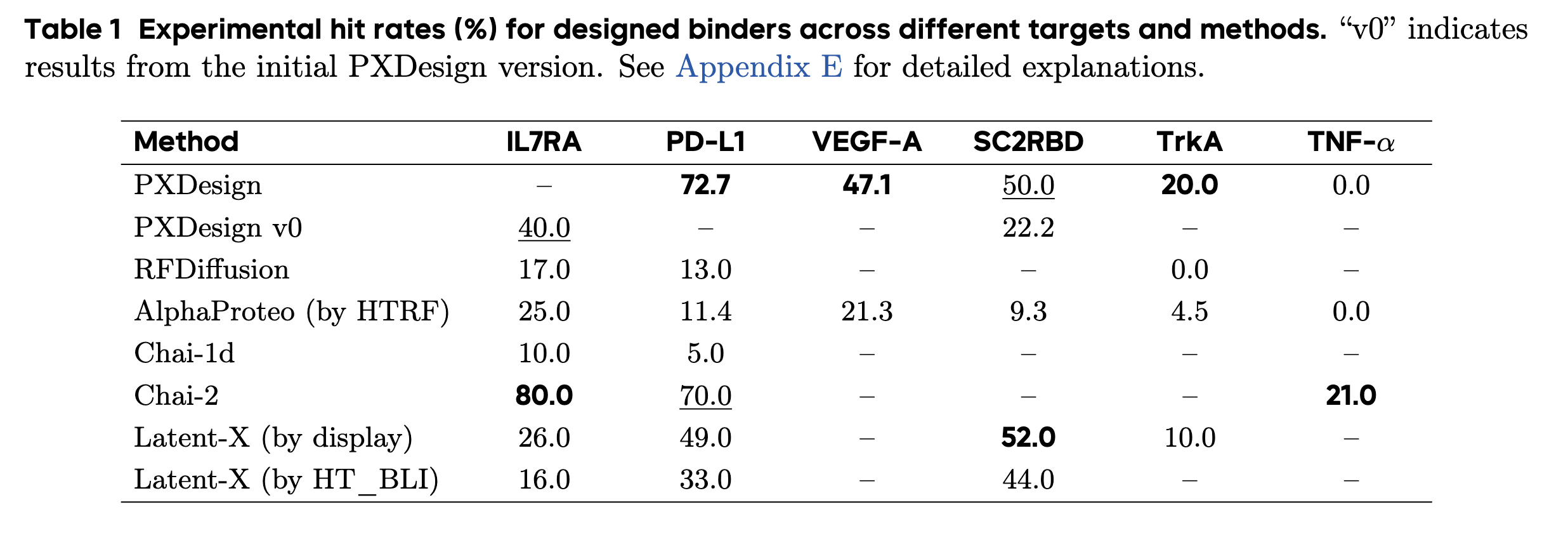
PXDesign claims impressive performance, comparable to Chai-2
Germinal
The Arc Institute has been on a tear for the past year or so, publishing all kinds of deep learning models, including the Evo 2 DNA language model and State virtual cell model.
Germinal is the latest model from the labs of Brian Hie and Xiaojing Gao, and this time they are joining in on the binder design fun. Installing this one was not easy, but eventually Claude and I got the right combination of jax, colabdesign, spackle and tape to make it run.
Unfortunately, there are also a couple of closed libraries required: IgLM, the antibody language model, and PyRosetta, which both require a license. AlphaFold 3 weights, which are thankfully optional, require you to petition DeepMind, but don't even try if you are a filthy commercial entity!
At some point all these tools need to follow Boltz and become fully open, or it will keep creating unnecessary friction and slowing everything down.
The code below uses Germinal to attempt one design for PD-L1. It should take around 5 minutes and cost <$1 to run (using a H100).
Note, I have not gotten Germinal to ever pass all its filters, which may be a bug, but it does still output designs with reasonable metrics.
The code was only released this week and is still in flux, so I don't recommend any serious use of Germinal until it settles down a bit.
My code below just barely works.
# Get the PD-L1 pdb from the Chai technical report
curl -O https://files.rcsb.org/download/5O45.pdb
# Make a yaml for Germinal
echo 'target_name: "5O45"\ntarget_pdb_path: "5O45.pdb"\ntarget_chain: "A"\nbinder_chain: "C"\ntarget_hotspots: "56,115,123"\ndimer: false\nlength: 129' > target_example.yaml
# Run Germinal; this is lightly tested, no guarantees of sensible output!
uvx --with PyYAML modal run modal_germinal.py --target-yaml target_example.yaml --max-trajectories 1 --max-passing-designs 1
Mosaic
Mosaic is a general protein design framework that is less plug-and-play than the others listed above, but enables the design of mini-binders, antibodies, or really any protein. It's essentially an interface to sequence optimization on top of three structure prediction models (AF2, Boltz, and Protenix.) You can construct an arbitrary loss function based on structural and sequence metrics, and let it optimize a sequence to that loss.
While mosaic is not specifically for antibodies, it can be configured to design only parts of proteins (e.g., CDRs), and it can easily incorporate antibody language models in its loss (AbLang is built in). The main author, Nick Boyd from Escalante Bio, wrote up a recent blog post on mosaic, and showed results comparable to the current state-of-the-art models like BindCraft. Unlike some other tools listed here, it is completely open.
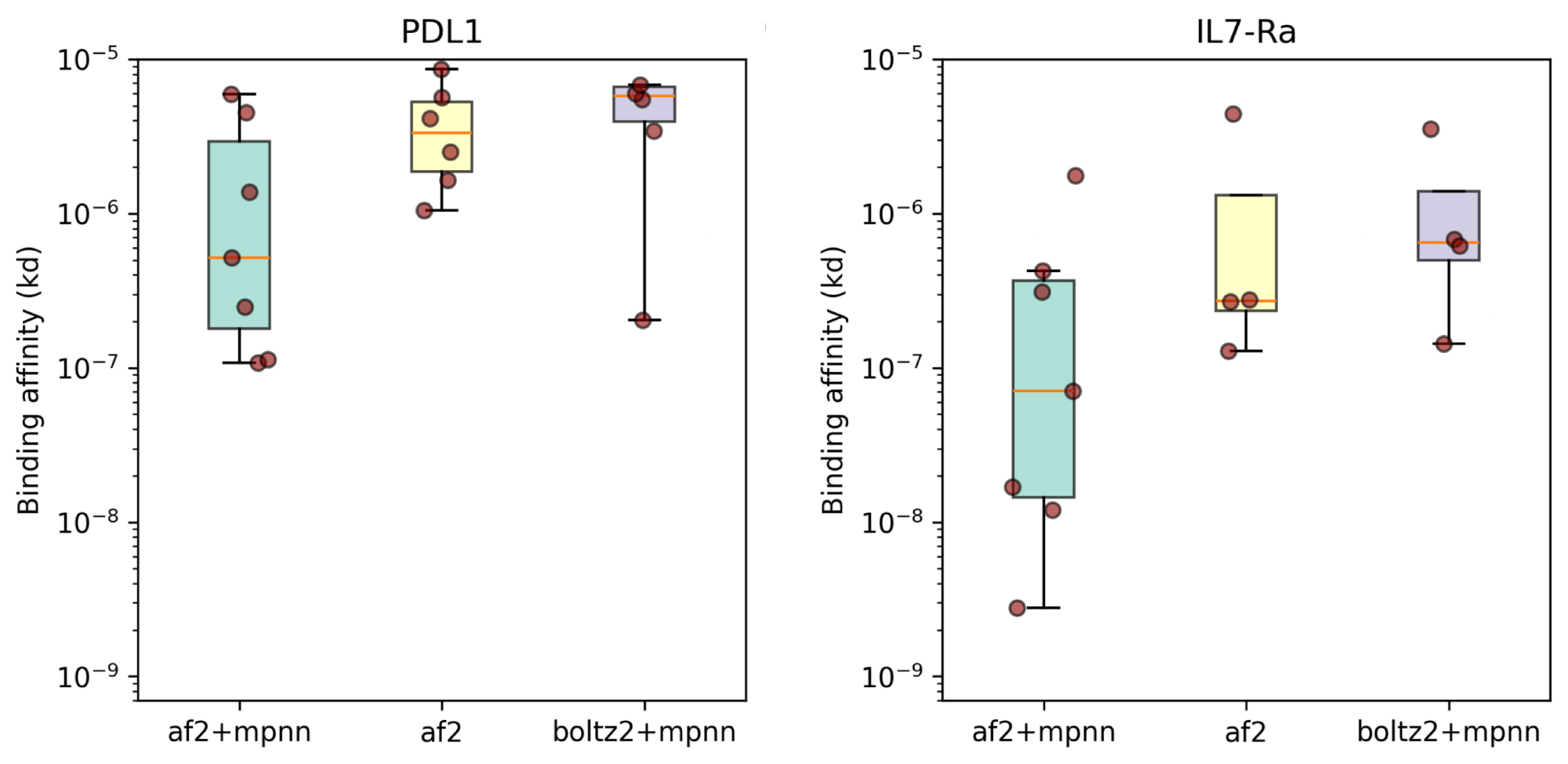
Mosaic has performance comparable to BindCraft on a small benchmark set (8/10 designs bound PD-L1 and 7/10 bound IL7Ra)
Commercial efforts
Chai-2
Chai-2 was unveiled in June 2025, and the technical report included some very impressive results. They claim a "100-fold" improvement over previous methods (I think this is a reference to RFantibody, which advised testing thousands of designs, versus tens for Chai-2.)
Chai-2 successfully created binding antibodies for 50% of targets tested, and some of these were even sub-nanomolar (i.e., potencies comparable to approved antibodies).
It is a bit dangerous to compare across approaches without a standardized benchmark—for example, some proteins like PD-L1 are easier to make binders for—but I think it's fair to say Chai-2 probably has the best overall performance stats of any model to date, mini-binder or antibody.
One criticism I have heard of these results is that the Chai team measured binding at 5-10uM in BLI,
which is not recommended as it can include weak binders.
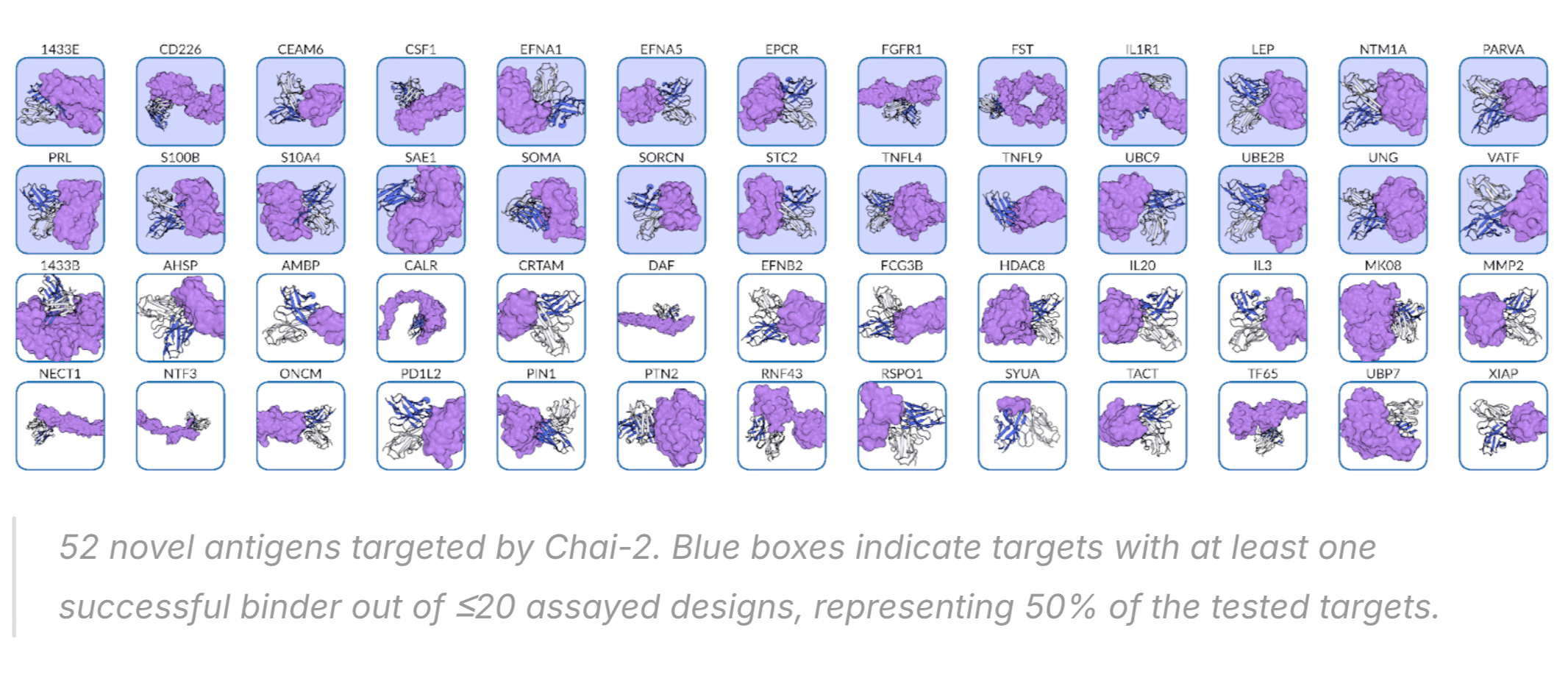
Nabla Bio
Like Chai, Nabla Bio appear to be focused on model licensing and partnering with pharma,
as opposed to their own drug programs.
This year they published a technical report on their JAM platform
where they demonstrated the ability to generate low-nanomolar binders against GPCRs,
a difficult but therapeutically important class of targets.
This may be one of very few examples where an AI approach has shown better performance than traditional methods,
rather than just faster results.
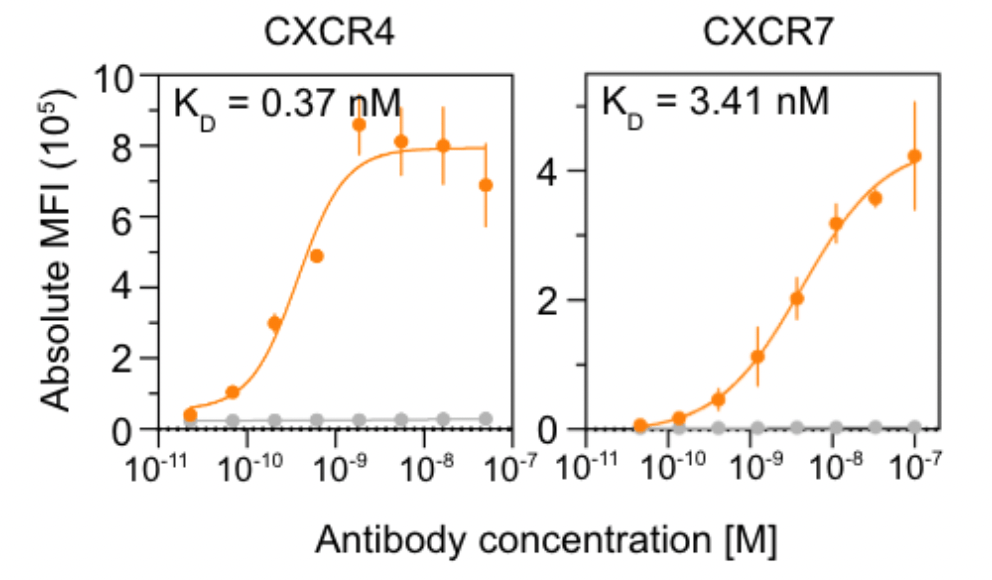
Nabla Bio showing impressive performance against two GPCRs
Diffuse Bio
Diffuse Bio's DSG2-mini model was also published in June 2025.
There is not too much information on performance apart from a claim that it "outperforms RFantibody on key metrics".
Like Chai-2, the Diffuse model is closed, though their sandbox is accessible so it's probably a bit easier to take for a test drive than Chai-2.
Screenshot from the Diffuse Bio sandbox
Tamarind, Ariax, Neurosnap, Rowan
Every year there are more online services that make running these tools easier for biologists.
Tamarind does not develop its own models, but allows anyone to easily run most of the open models.
Tamarind have been impressively fast at getting models onboarded and available for use.
They have a free tier, but realistically you need a subscription to do any real work, and I believe that costs tens of thousands per year.
Neurosnap looks like it has similar capabilities to Tamarind, but the pricing may be more suitable for academics or more casual users.
Ariax has done an incredible job making BindCraft (and FreeBindCraft) available and super easy to run.
They don't generate antibodies yet, but they will once a suitably open model is released.
Rowan is more small molecule- and MD-focused than antibody-focused—they even release their own MD models—so although a fantastic toolkit, less relevant to antibody design.
Tamarind has over one hundred models, including all the major structure prediction and design models
Xaira, Generate, Cradle, Profluent, Isomorphic, BigHat, etc
There are a gaggle of other actual drug companies working on computational antibody design, but these models will likely stay internal to those companies. Cradle is the outlier in this list since it is a service business, but I believe they do partnerships with pharma/biotech, rather than licensing their models.
It will be interesting to see which of these companies figure out a unique approach to drug discovery, and which get overtaken by open source.
Most people in biotech will tell you that if you want a highly optimized antibody and can wait a few months, companies like Adimab, Alloy, or Specifica can already reliably achieve that, and the price will be a small fraction of the total cost of the program anyway.
Benchmarks
AIntibody
The AIntibody competition, run by the antibody discovery company Specifica, is similar to last year's Adaptyv binder design competition, but focused on antibodies.
The competition includes three challenges, but unlike the Adaptyv competition, none of the challenges is a simple "design a novel antibody for this target".
The techniques used in this competition ended up being quite complex workflows specific to the challenges: for example, a protein language model combined with a model fine-tuned on affinity data provided by Specifica.
Interestingly, the "AI Biotech" listed as coming third is—according to their github—IgGM. The Specifica team has given a webinar on the results with some interesting conclusions, but the full write-up is still to come.
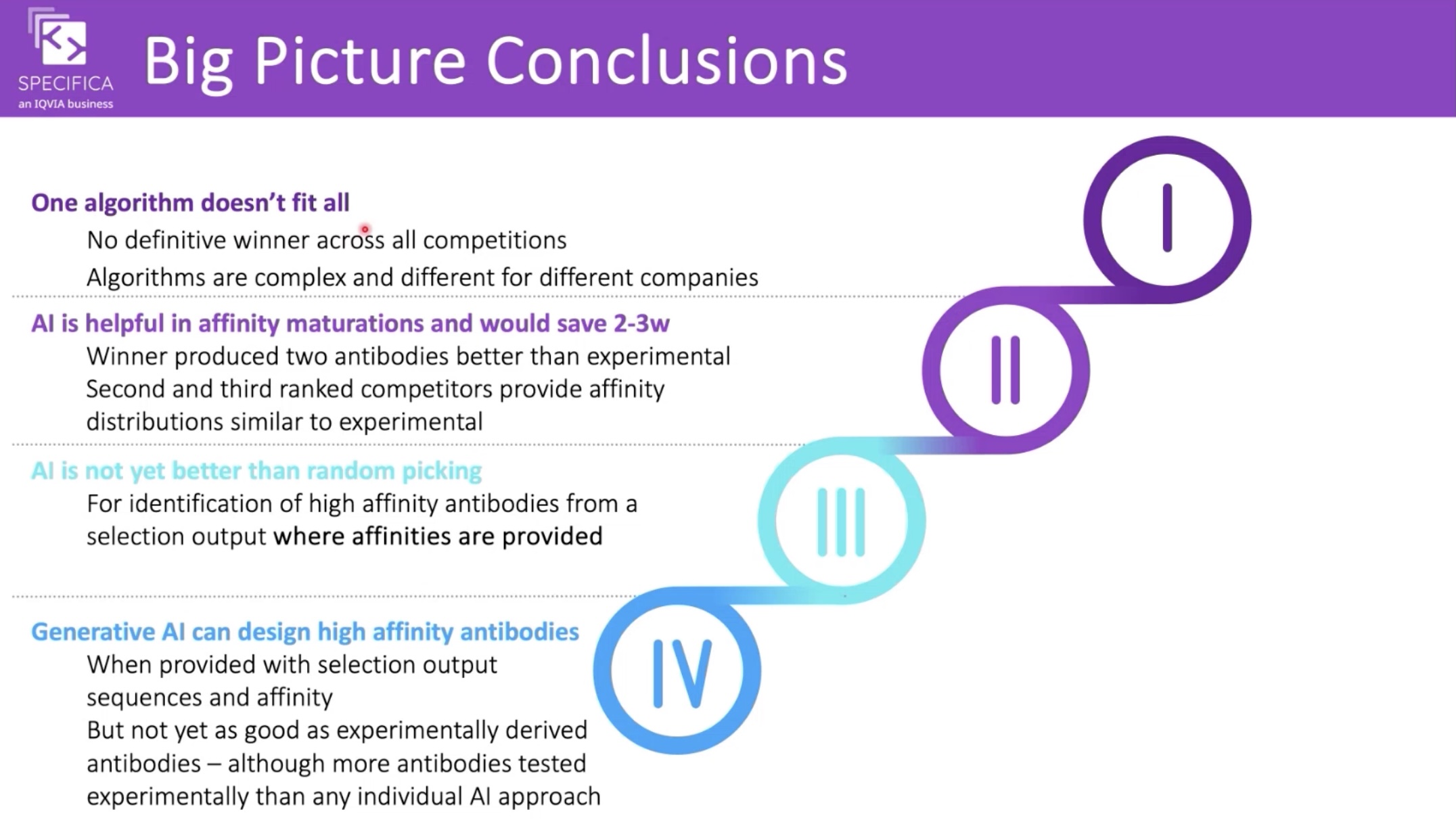
Conclusions from the AIntibody webinar
Ginkgo
Just this week,
Ginkgo Datapoints
launched a kaggle-style
competition on huggingface
with a public leaderboard.
This challenge is to predict developability properties (expression, stability, aggregation),
which is a vital step downstream of making a binder.
The competition deadline is November 1st.
BenchBB
BenchBB is Adaptyv Bio's new binder design benchmark.
While it's not specifically for antibodies,
if you did try to generate PD-L1 binders using the biomodals commands given above,
you could test your designs here for $99 each.
We know we need a lot more affinity data to improve our antibody models,
and $99 is a phenomenal deal, so some crypto science thing should fund this instead of the nonsense they normally fund!

There are seven currently available BenchBB targets
Conclusion
I often seem to end these posts by saying things are getting pretty exciting. I think that's true, especially over the past few weeks with IgGM and Germinal being released, but there are also some gaps. RFantibody was published quite a while ago, and we still only have a few successors, most of which are not fully open. The models are improving, but large companies like Google (Isomorphic) are no longer releasing models, so progress has slowed somewhat. Mirroring the LLM world, it's left to academic labs like Martin Pacesa, Sergey Ovchinnikov, Bruno Correia and Brian Hie, and Chinese companies like Tencent to push the open models forward.
I did not talk about antibody language models here even though there are a lot of interesting ones. It would be a big topic, and they are more applicable to downstream tasks, once you have a binder to improve upon.
As with protein folding (see SimpleFold from this week!), there is not a ton of magic here, and many of the methods are converging on the same performance, governed by the available data. To improve upon that, we/someone probably needs to spend a few million dollars generating consistent binding and affinity data. In my opinion, Adaptyv Bio's BenchBB is a good place to focus efforts.
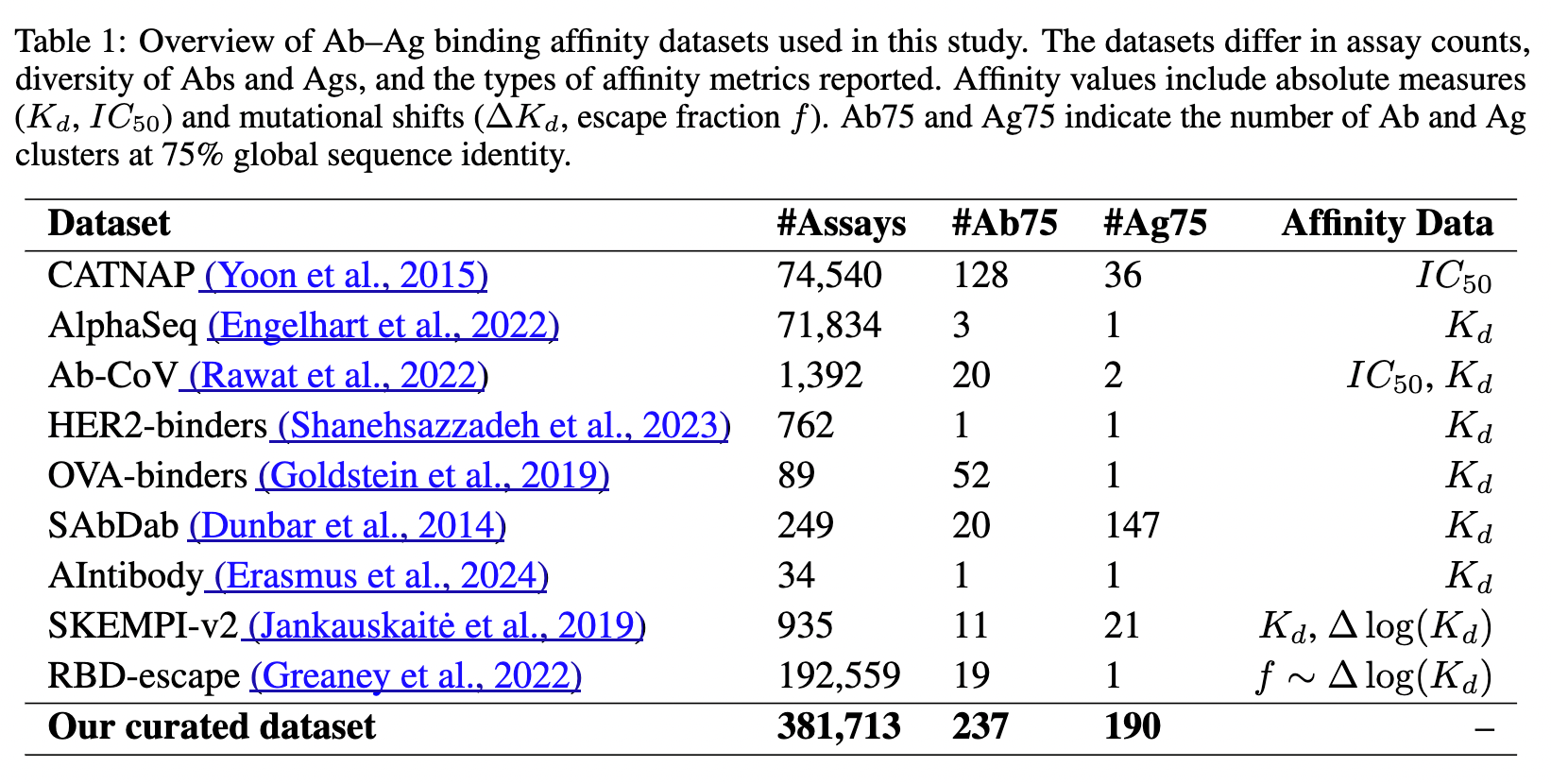
Publicly available affinity data from the AbRank paper. Most of the data is from SARS-CoV-2 or HIV, so it's not nearly as much as it seems.
Running the code
If you want to run the biomodals code above and design some antibodies for PD-L1 (or any target) you'll need to do a couple of things.
1. Sign up for modal.
They give you $30 a month on the free tier, more than enough to generate a few binders.
2. Install uv. If you use Python you should do this anyway!
3. Clone my biomodals repo:
git clone https://github.com/hgbrian/biomodals # or gh repo clone hgbrian/biomodals