The new class of protein AI design tools are amazing, and could revolutionize many areas of science, including therapeutics, diagnostics, and biosensors. Surprisingly, one important area that I haven't seen discussed too much is how these tools could impact patents. I am not a lawyer, so obviously this post is just my basic understanding, and I'd be happy to hear corrections. If there is a more expert critique I did not find it.
Patents are wordy and convoluted by design. For proteins, because a string of amino acids defines them, there are some common elements: they often include the sequence(s) being patented, and a threshold for how similar another sequence can be before infringing. That means there is a target to hit, and AI is really good at hitting targets.
There are two major categories of protein patents: biologics (usually meaning antibodies) and enzymes.
Antibodies
According to the European Patent Office, there are two main ways to patent an antibody:
- "functional" claims, usually meaning the antibody's associated antigen or epitope;
- "structural" claims, usually meaning a sequence and sequence identity threshold, along with the epitope or some other support.
Over the past few years, the "functional" claim has been going away. In the US it was killed off by the 2023 Amgen vs Sanofi ruling, which essentially said you can't patent the concept of an antibody against PCSK9. That means antibodies are now almost exclusively patented based on their structure (more specifically, a sequence plus some supporting functional information like epitope affinity.)
For antibody sequences, it used to be common for claims to cover any sequence 80%+ identical in the heavy or light chains. These days it seems like you have to be more specific, with claims only covering 100% identity to all 6 CDRs.
To take some real examples:
- Zanidatamab, a HER2 bispecific approved in 2024, claims sequences with 100% sequence identity to its CDRs;
- Epcoritamab, a CD3/CD20 bispecific approved in 2024, also has claims sequences with 100% sequence identity to its CDRs;
- Trastuzumab, the famous HER2 antibody approved in 1998 (filed in 2013), claims sequences with 85%+ sequence identity to the heavy and light chains, and does not mention CDRs at all.
The EPO says: "the slightest modification of the CDRs can affect the recognition of the target." There is a nice breakdown of the differences between the USPTO vs EPO approach to antibody patents here.
Enzymes
For enzymes, the patent landscape is more complicated, or at least more varied. Unlike antibodies, where the patents are pretty uniformly focused on the sequence that binds an epitope, enzymes can perform any number of functions. Enzyme types include enzyme replacement therapies, industrial enzymes like detergents, and molecular biology tools like CRISPR-Cas9. It is still typical for these patents to include a sequence and supporting information.
Some examples:
- this detergent patent, granted in 2018, claims sequences with 60%+ sequence identity to the reference;
- this proteinase patent, granted in 2022, claims sequences with 90%+ sequence identity to the reference;
- this novel Taq polymerase patent, granted in 2025, claims sequences with 95%+ sequence identity to the reference.
Cas9
The Cas9 patents are unusually diverse: there are hundreds of them and they mostly cover the many applications of the invention rather than the sequences. Since the 2013 ruling against Myriad Genetics, sequences from naturally occurring enzymes like Cas9 cannot be patented. Engineered sequences can be patented with other supporting functional information. You cannot take one of the thousands of unique Cas9 sequences in GenBank and use that to circumvent the CRISPR-Cas9 patents.
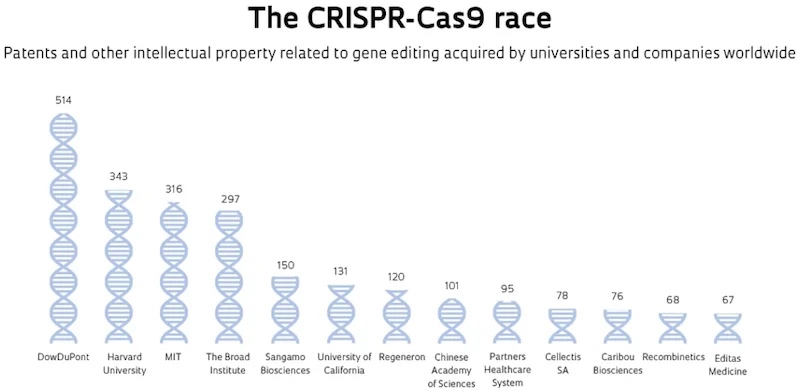
There are hundreds of Cas9 patents covering everything anyone could think of
AI
Given that the amino acid sequence is so important in protein patents, I am surprised that it is not bigger news that AI has effectively broken the direct connection between sequence and function.
For patents where protein sequence identity is protected, it is now relatively straightforward to generate new sequences that fold to the same structure but have 50% or lower sequence identity.
For antibody patents where the CDR sequence is protected, I believe it is also relatively straightforward to introduce a mutation that does not disrupt binding. To be honest, I am not even sure AI is required here, since a mutation scan could perform the same function. Perhaps for this reason, a recent paper called for "comprehensive CDR scanning" to protect a panel of CDR sequences instead of just one.
ProteinMPNN, published in 2022 by Baker lab, is the most prominent tool for producing a new sequence that folds to a known structure. ProteinMPNN is widely used as a step in many protein design workflows. For example, methods like RFdiffusion generate backbone coordinates only, and ProteinMPNN turns that into an amino acid sequence.
In a follow-up ProteinMPNN paper, the authors demonstrated that they could make a myoglobin and TEV protease with comparable or better function and greater stability than the natural versions, with sequence identities as low as 40%. This is below the sequence identity threshold in any patent I have seen.
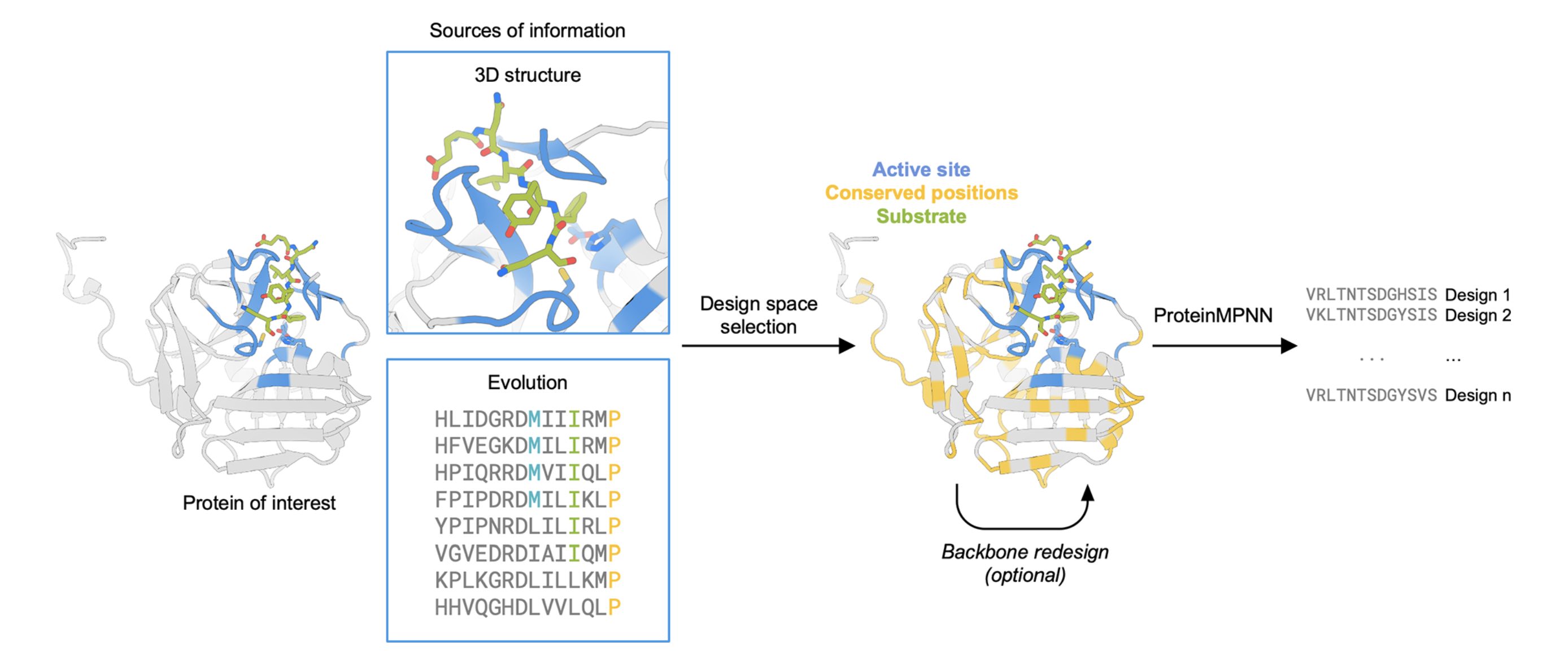
ProteinMPNN can be used to produce a new sequence for a protein while maintaining its function
Sequence vs Structure
If this ability for AI to circumvent sequence-based patents is an issue, maybe the obvious change here would be to base patent protection on structure. This is a bit more complex than sequence identity, but one way to do this would be with TM-align or a similar tool. TM-align has >3k citations so it is arguably the standard in the field. A TM-score of above 0.8 indicates "the same topology"—in other words a very close structure. I think this would work well for many proteins, though it might need to be constrained to subdomains (akin to CDRs) in some cases.
Interestingly, the only literature I found on patenting 3D structure is from 20 years ago. Maybe this has been debated already and rejected for some reason. I suspect it was just easier to use sequence though.
OpenCRISPR-1
OpenCRISPR-1 was published in 2024 by the protein AI company Profluent. This is a de novo Cas9 enzyme that is substantially different in sequence to any known Cas9 (according to the abstract, "400 mutations away in sequence [from SpCas9]"—specifically 403/1380, or 71% identity).
Cas9 is a bilobed enzyme, with a REC lobe (nucleotide recognition) and a NUC lobe (DNA cleavage and PAM recognition.) Broadly speaking, the REC lobe is the first half of the enzyme (amino acids 50–700), and the NUC lobe is the second (1–50 and 700–1350.) These two lobes are connected by a "bridge helix".
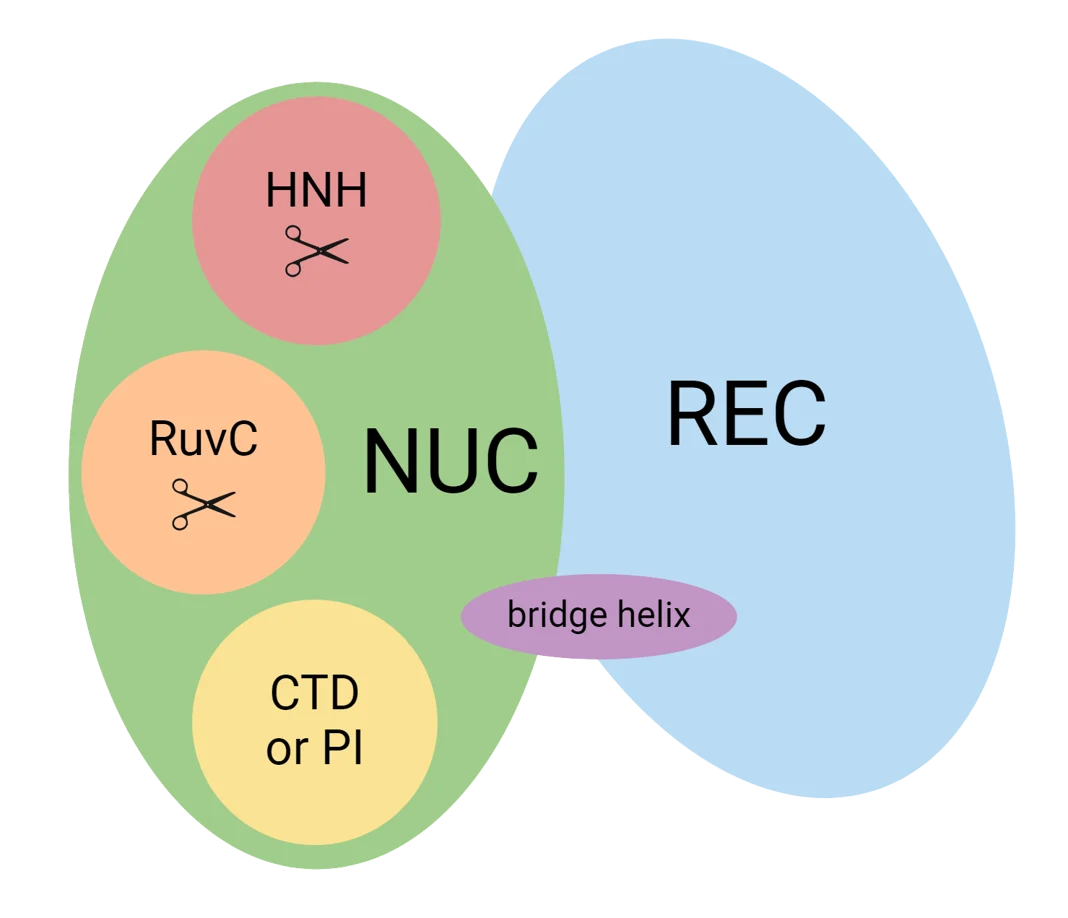
Cartoon representation of Cas9 from addgene.
The OpenCRISPR-1 enzyme is not as novel as it might seem. In fact, I found it is actually 98% identical to a sequence constructed from three Cas9's spliced together from Streptococcus cristatus, Streptococcus pyogenes and Streptococcus sanguinis (24 amino acids are unique to OpenCRISPR-1).
This raises an interesting question, which is whether you could create a "novel" Cas9 by simply stitching together the REC lobe from one species' Cas9 and the NUC lobe from another. I believe this enzyme would work, and this sequence would meet any sequence identity threshold requirements.
The Profluent paper says the OpenCRISPR-1 enzyme was released for "research and commercial applications", but there is a big caveat here. Since CRISPR-Cas9 patents post-date the Myriad decision, almost all are functional / method of use, and naturally the most protected part is the use of Cas9 in "commercial applications" like therapeutics and diagnostics.
It is commendable that Profluent tried to broaden the availability of Cas9, so I appreciate the work behind this, but as I understand it, OpenCRISPR-1 is not really more available for commercial use than any Cas9.
There is actually another "royalty-free" Cas, a "Class 2 Type V" Cas nuclease called MAD7, released by Inscripta for commercial use in 2023. I do not know how this enzyme intersects with the many Cas9 patents.
Conclusion
One upshot of all this AI work is that me-too and biosimilar antibodies will be easier to make. That saves some time and money, but does not necessarily save on the major clinical trial costs, although the probability of success could go up a lot if the antibody is functionally identical.
While many enzyme patents will be affected, patents like CRISPR-Cas9 that rely on functional or method of use claims do not seem to be impacted as much. I don't know how many enzyme patents rely on sequence identity claims vs other claims these days. It would be interesting to (get an AI to) do a proper survey.
For internal research use, it's unclear to me that using AI to reproduce a patented protein does a whole lot, since at least in drug development, the research exemption seems to allow for the use of patented material quite broadly.