Recently, a friend recommended BioGaia Prodentis to me. It is a DTC oral probiotic you can buy online that is supposedly good for oral health. I thought it would be interesting to do some sequencing to see what, if anything, it did to my oral microbiome.

BioGaia Prodentis is available online for $20 or less for a month's supply
BioGaia
BioGaia has a fascinating story. They are a Swedish company, founded over 30 years ago, that exclusively sells probiotics DTC. They have developed multiple strains of Limosilactobacillus reuteri, mainly for gut and oral health. They apparently sell well! Their market cap is around $1B—impressive for a consumer biotech.
Going in, I expected scant evidence for any real benefits to their probiotics, but the data (over 250 clinical studies) is much more complete than I expected.
Most notably, their gut probiotic, Protectis, seems to have a significant effect on preventing Necrotizing Enterocolitis (NEC) in premature babies. According to their website:
5-10% of the smallest premature infants develop NEC, a potentially lethal disorder in which portions of the bowel undergo tissue death.
In March 2025, the FDA granted Breakthrough Therapy Designation to IBP-9414, an L. reuteri probiotic developed by BioGaia spinout IBT.
This is not specifically for the oral health product, but it's for sure more science than I expected to see going in.
Prodentis
BioGaia Prodentis contains two strains of L. reuteri: DSM 17938 and ATCC PTA 5289. The claimed benefits include fresher breath, healthier gums, and outcompeting harmful bacteria.
Sequencing with Plasmidsaurus
Many readers will be familiar with Plasmidsaurus. Founded in 2021, the team took a relatively simple idea: use massively multiplexed Oxford Nanopore (ONT) to offer complete plasmid sequencing with one day turnaround for $15, and scaled it. Plasmidsaurus quickly became part of biotech's core infrastructure, and spread like wildfire. It also inspired multiple copycats.
Compared to Illumina, ONT is faster and has much longer reads, but lower throughput and lower accuracy. This profile is a great fit to many sequencing problems like plasmid QC, where you only need megabases of sequences, but want an answer within 24 hours.
Over time, Plasmidsaurus has been adding services, including genome sequencing, RNA-Seq, and microbiome sequencing, all based on ONT sequencing.
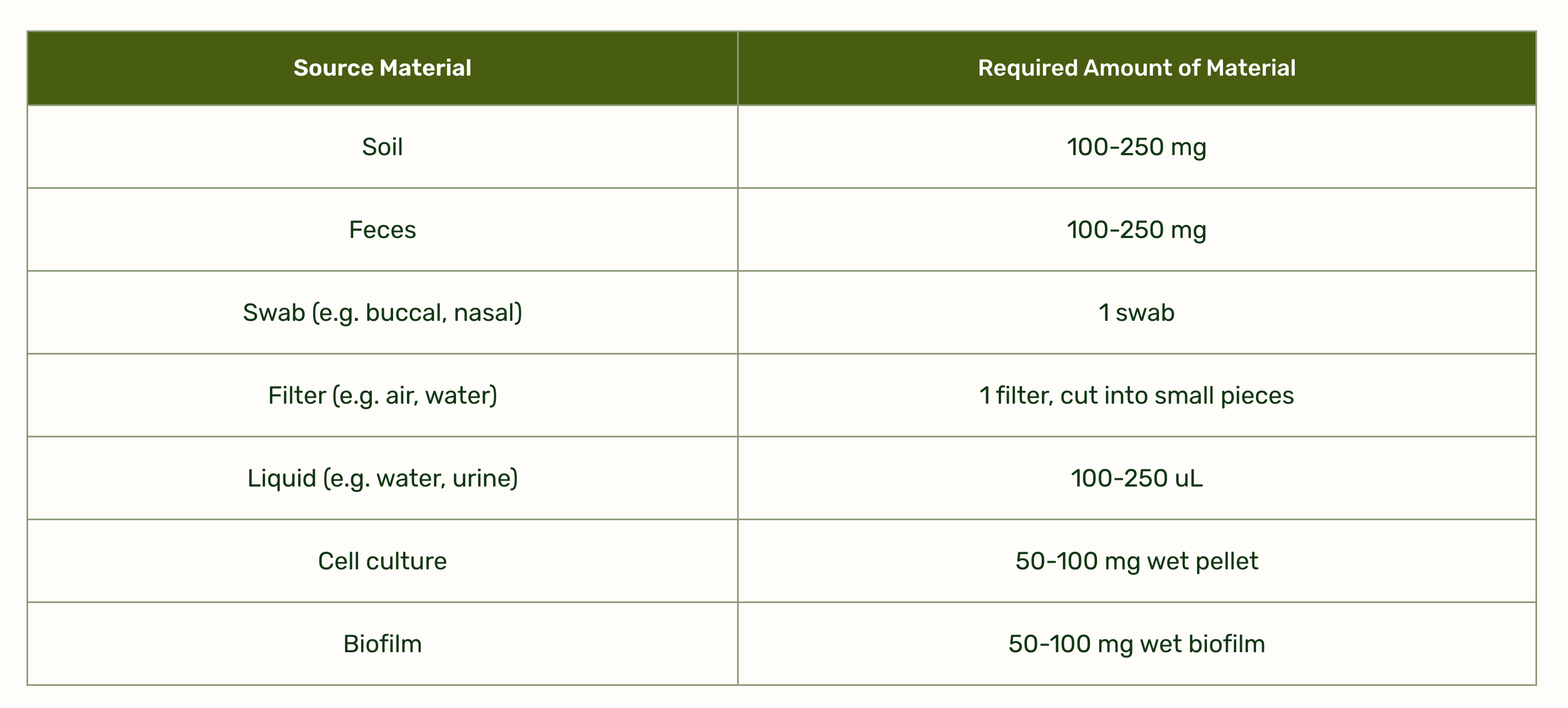
Plasmidsaurus accepts many kinds of sample for microbiome sequencing
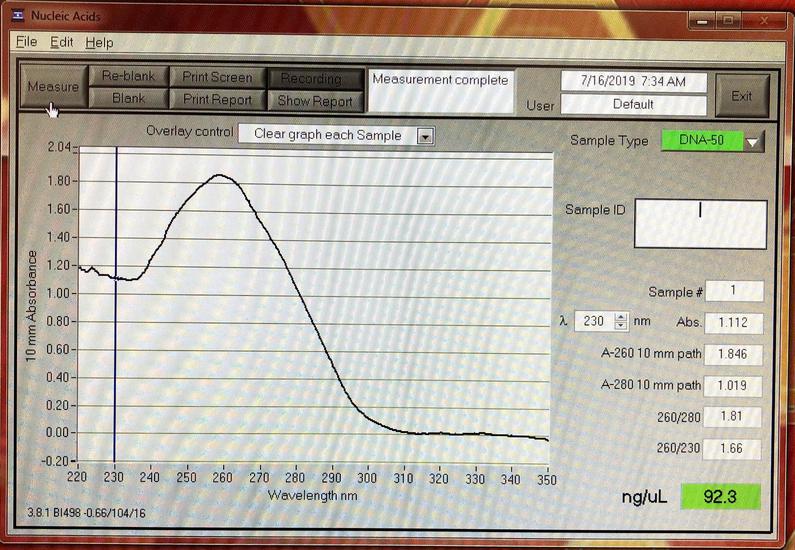
I used their 16S sequencing product, which costs $45 for ~5000 reads, plus $15 for DNA extraction. 16S sequencing is an efficient way to amplify and sequence a small amount of DNA (the ubiquitous 16S region) and be able to assign reads to specific species or even strains.
This experiment cost me $240 for four samples, and I got data back in around a week. It's very convenient that I no longer have to do my own sequencing. As a side note, you can pay more for more than 5000 reads, but unless you want information on very rare strains (<<1% frequency), this is a waste of money.
Sample collection is simple: take 100-250 µL of saliva and mix with 500 µL of Zymo DNA/RNA Shield (which I also had to buy for around $70.) You also need 2 mL screwtop tubes to ship in.
The reads are high quality for nanopore sequencing, with a median Q score of 23 (99%+ accuracy). This is more than sufficient accuracy for this experiment. The read length is very tightly distributed around 1500 nt (the length of a typical 16S region).
The results provided by Plasmidsaurus include taxonomy tables, per-read assignments, and some basic plots. I include a download of the results at the end of this article, as well as the FASTQ files.
The experiment
The main idea of the experiment was to see if any L. reuteri would colonize by the end of 30 days of probiotic use, and if so, whether it would persist beyond that. I collected four saliva samples:
| Sample | Timing | Description |
|---|---|---|
| Baseline A | Day -4 | A few days before starting BioGaia |
| Baseline B | Day -1 | The day before I started BioGaia |
| Day 30 | Day 30 | The last day of the 30 day course |
| Week after | Day 37 | One week after completing the course |
Heatmap of the top 20 species. All species assignments were done by Plasmidsaurus
Did L. reuteri colonize?
There was no L. reuteri found in any of the samples. I did a manual analysis to check for any possible misassignments, but the closest read was only 91% identical to either L. reuteri strain.
The probiotic either (a) didn't colonize the oral cavity; (b) was present only transiently while actively taking the lozenges; (c) was below the detection threshold.
Probiotics are generally bad at colonizing, which is why you have to keep taking them. Still, I was surprised not to see a single L. reuteri read in there.
What actually changed?
Even though the probiotic itself didn't show up, the oral microbiome did change quite a lot.
The most striking change was a massive increase in S. salivarius. S. salivarius went from essentially absent to ~20% of my oral microbiome on the last day. However, this happened one week after I stopped taking the probiotic, so it's very unclear if it is related.
| Sample | S. mitis | S. salivarius |
|---|---|---|
| Baseline A | 2.0% | 0.4% |
| Baseline B | 15.9% | 0.0% |
| Day 30 | 10.2% | 0.8% |
| Week after | 1.0% | 19.3% |
We see S. mitis decreasing as S. salivarius increases, while the total Streptococcus fraction stayed roughly stable. It's possible one species replaced the other within the same ecological niche.
S. salivarius is itself a probiotic species. The strain BLIS K12 was isolated from a healthy New Zealand child and is sold commercially for oral health. It produces bacteriocins that kill Streptococcus pyogenes (strep throat bacteria).
At the same time, V. tobetsuensis increased in abundance from 2.1% to 5.7%. Veillonella bacteria can't eat sugar directly—they survive by consuming lactate that Streptococcus produces. The S. salivarius bloom is plausibly feeding them.
Are these changes real or intra-day variation?
There was a lot more variation in species than I expected, especially comparing the two baseline samples. In retrospect, I should have taken multiple samples on the same day, and mixed them to smooth it out.
However, there is some light evidence that the variation I see is not just intra-day variation. Specifically, there are several species that stay consistent in frequency across all samples: e.g., Neisseria subflava, Streptococcus viridans, Streptococcus oralis.
Conclusions
- L. reuteri didn't detectably colonize my mouth. Oral probiotics are surprisingly difficult to detect, even if you sample the same day as dosing.
- S. salivarius increased massively in abundance, but this increase happened after I stopped taking BioGaia
- Microbiome sequencing can be used to assess oral health. None of the "red complex" bacteria (P. gingivalis, T. forsythia, T. denticola) associated with gum disease were found in any sample.
- The oral microbiome is dynamic, with huge swings in species abundance over a timeframe of just weeks
- Microbiome sequencing is very easy and convenient these days thanks to Plasmidsaurus
- Prodentis tastes good, may help with oral health, and I'd consider taking it again