In the past, I have had trouble extracting high quality DNA for sequencing, so I decided I need a spectrophotometer to help figure this out — or at least to stop running poor quality samples.
In this post I'll report what I learned about spectrophotometers and what happens when I test a few water samples with one.
DNA quantitation
It seems like the terms spectrophotometer and spectrometer are essentially synonymous. Spectrophotometers are generally focused on absorption (e.g., DNA absorbing at 260nm) and spectrometers are focused on emission (e.g., stars emitting light at various wavelengths).
One of the major uses of spectrophotometers in the lab is DNA quantitation. Despite that, most regular spectrophotometers are unsuitable for this purpose. Typically, you want readings at 230nm (sugars, salts, organic solvents), 260nm (nucleic acid), 280nm (protein), and 320nm ("turbidity", air bubbles, other stuff). The most often used metrics for DNA purity are the 260/280 and 260/230 ratios. Notably, none of these wavelengths are in the visible spectrum.
There are a few ways you can measure DNA quantity in solution:
- measure absorption at all wavelengths from ~200 to 400nm and plot a curve
- measure absorption at the specific wavelengths listed above
- add a dye that fluoresces upon binding nucleic acid and measure that wavelength
DIY Spectrometers
The problem doesn't sound too complicated, so you'd think there might be a way to do this cheaply. Once you can detect the appropriate wavelengths, the rest is simple.
In fact, there are several DIY spectrometer projects out there, including a nice Public Lab one. (I bought that one a few years ago and it was a fun toy.) Even though at least one of these projects describes itself as a DIY Nanodrop, I think this is a misleading name. The volume required is low, but like all of these projects, it uses a regular LED and camera, so it only measures light in the visible spectrum (~400-750nm, plus some IR and a little UV).
Visual spectrum spectrophotometers are fine for OD readings (which uses 600nm, aka yellow), but that's about it.
Another DIY option you could imagine is to use a UV lamp and bandpass filters like these from Edmunds Optical, and a simple photosensor. It's actually not even easy to get a UV lamp that emits at 230nm, and the optical filters are surprisingly expensive ($300 each), so sadly this is not an economical option.
Nanodrop vs Qubit
The most popular lab tools for DNA quantitation are the Nanodrop and the Qubit. These devices are prohibitively expensive: a Nanodrop is about $7k new and $3k+ second-hand, and a Qubit is $3k and $1k+, respectively.
The main difference between the two is that the Qubit is a fluorometer, and hence requires a nucleic-acid–binding dye. This makes the Qubit more accurate for quantitation, since other molecules absorb at 260nm, but also a bit more work. The Nanodrop has the advantages of using very little material (~1µl), and the ability to detect contaminants at other wavelengths. Either may suit, depending on whether quantitation or contamination is more important. A recent Twitter thread covered this topic; apparently every other lab calls a nanodrop a "random number generator".

GeneQuant
I thought there might be a sweet spot device that measures only at 230/260/280/320nm. There was even a promising one on alibaba a while ago, that sadly turned out not to be real... It turned out there was a better option: I learned about the GeneQuant, the device I ended up buying, from this relevant Google Groups thread.
I bought a second-hand GeneQuant (25 years old!) on ebay for ~$150. The device I bought came with a nice 500µl cuvette (the sample holder you insert), which is nice, but an enormous volume for DNA quantitation. Unfortunately, cuvettes are expensive, I assume due to the high quality quartz required, and the lower the cuvette volume, the more they cost. I'll probably need a 10µl cuvette at some point.
Even though this GeneQuant is old, I thought it would probably work fine for my purposes, because even if it's miscalibrated, I should still be able to learn what a contaminated sample looks like.

Water Purity
At home we use a PUR filter, which is supposedly better than a Brita, at least according to The Wirecutter. I have been periodically curious if it is doing much, since our tap water is pretty good — especially towards the end of the filter's life when the filtration slows down.
This is a useful experiment because it's also a good way to check the precision and reproducibility of readings from the GeneQuant.
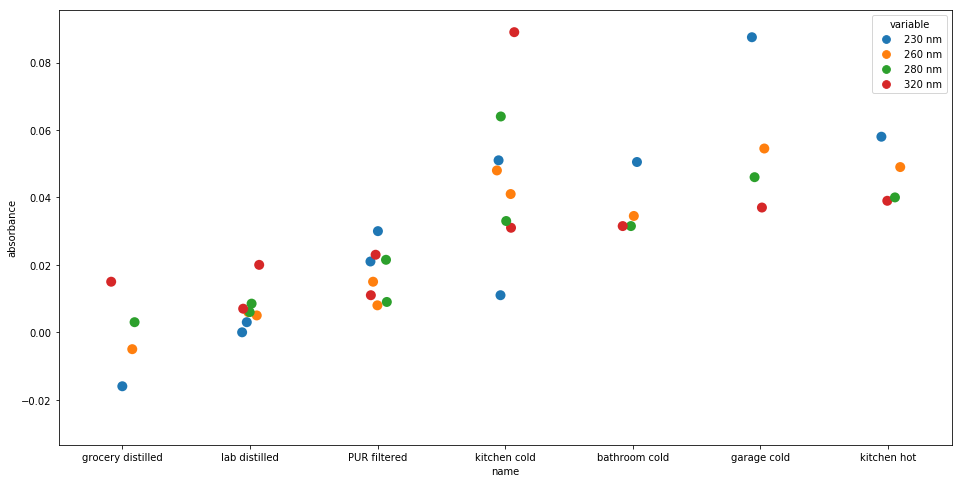
I took samples from the kitchen sink, bathroom sinks, PUR filter, distilled water bought at a grocery store, and nuclease-free lab-grade water.
import numpy as np
import pandas as pd
from io import StringIO
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv(StringIO("""name,sample,230 nm,260 nm,280 nm,320 nm
kitchen cold, 2020-05-03a, 0.011, 0.048, 0.064, 0.089
lab distilled, 2020-05-03a, 0.003, 0.005, 0.006, 0.007
PUR filtered, 2020-05-03a, 0.021, 0.008, 0.009, 0.011
kitchen cold, 2020-06-03a, 0.051, 0.041, 0.033, 0.031
kitchen cold, 2020-06-03a, 0.051, 0.041, 0.033, 0.031
PUR filtered, 2020-06-03a, 0.027, 0.014, 0.019, 0.023
PUR filtered, 2020-06-03a, 0.033, 0.016, 0.024, 0.023
kitchen hot, 2020-06-03a, 0.056, 0.047, 0.038, 0.037
kitchen hot, 2020-06-03a, 0.060, 0.051, 0.042, 0.041
lab distilled, 2020-06-03a, 0.005, 0.011, 0.014, 0.025
lab distilled, 2020-06-03a,-0.005, 0.001, 0.003, 0.015
grocery distilled,2020-06-03a,-0.016,-0.005, 0.002, 0.015
grocery distilled,2020-06-03a,-0.016,-0.005, 0.003, 0.016
grocery distilled,2020-06-03a,-0.016,-0.005, 0.003, 0.015
bathroom cold, 2020-06-03a, 0.051, 0.035, 0.032, 0.032
bathroom cold, 2020-06-03a, 0.050, 0.034, 0.031, 0.031
garage cold, 2020-06-03a, 0.088, 0.055, 0.046, 0.037
garage cold, 2020-06-03a, 0.087, 0.054, 0.046, 0.037
"""))
df
df_plot = (
df.groupby(['name', 'sample'])
.agg('median')
.reset_index()
.drop('sample', axis=1)
.melt(id_vars=["name"])
.rename(columns={"value":"absorbance"})
)
f, ax = plt.subplots(figsize=(16,8))
sns.stripplot(data=df_plot.sort_values("absorbance"),
x="name",
y="absorbance",
hue="variable",
size=10);
Conclusions
I didn't bother to run many replicates, but still, there are some pretty interesting results here:
- the precision of the instrument is very high — at least when readings are taken close together — which is encouraging.
- the distilled water from the grocery store is "cleaner" than the lab-grade water. There are confounders, like the fact that the lab-grade water is older, but it's still interesting to see the grocery store water coming back so pure.
- The PUR is basically half-way between distilled water and tap water, so it's definitely doing something. It's unclear to me if the residual difference is stuff you want (minerals etc. that influence the taste) or just incomplete purification.
- The various faucets in the house produce similar purity water, as does hot and cold water. I thought faucets that had not been used in many days, like the garage sink, might have some detectable residue, but if it's there, it's a minor difference at most.
Another interesting thing I recently noticed about tap water vs distilled is that when we ran a humidifier with tap water, eventually the PM2.5 in the house would climb way beyond 10µg/m3 (which is very bad). I thought this might be just aerosolized water, but it didn't happen with distilled water. I don't know which impurities were causing this issue, so it could be harmless, but we completely stopped using tap water in the humidifier after that.