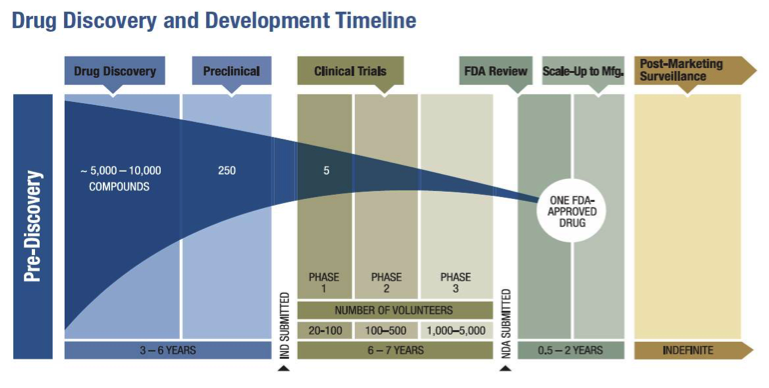
I looked around for a broad review of this area, discussing classes of drug targets and therapies, but didn't really find anything. These are some notes I made in an effort to help understand the landscape.
Drug target types
Every drug has a "target", a biomolecule that the drug affects in humans. There are only a few options for targets.
-
Underexpressed protein
Sometimes, a protein is lacking and needs to be upregulated or replaced. The solution, for extracellular proteins, can be as simple as injecting the missing protein. This may sound like it's an obscure element of drug development, but it's basically the foundation of biotech. Genzyme started out replacing the missing enzymes in Gaucher and Fabry disease; Genentech got their start with recombinant insulin, an underexpressed peptide; Amgen's first big drug was recombinant G-CSF (neupogen). -
Overexpressed protein
Sometimes, you are making too much protein and you need to dial it down. Many cancer targets are like this (e.g., HER2 amplification), though some are mutated too. This is one of the easiest scenarios for drug development, because all you have to do is knock the protein down with high specificity. It's much easier to break something than fix it. -
Malfunctioning protein
If you have a genetic disease, there's a good chance it's due to an absent or malfunctioning protein. Cystic fibrosis has over a thousand known causative mutations and most of these mutations cause misfolding. How do you fix a misfolded protein? It's always difficult and sometimes impossible. Vertex's CF drugs represent one unusual success story. Their latest drug, Orkambi, is actually two drugs in one: one that increases CFTR's activity (Kalydeco, a drug originally for the G551D mutation only), and the other helps the mutated CFTR fold properly. -
Non-proteins
There are things in the body besides proteins, for example, hormones, lipids, DNA. These are much less common targets for therapies, though amazingly gene therapy is starting to become feasible. (There are now two human gene therapies approved in Europe, and several veterinary gene therapies). According to a 2016 review in NRDD, FDA-approved drugs targeted non-proteins in only 28/695 cases.
Choice of Therapy
There are a plethora of options these days.
- Small molecules
- Chemical
This is what most people mean by a drug. You use chemistry to make them: perhaps you have a large library of randomly generated chemicals that you screen against a target, or perhaps you design your drug and simulate target binding computationally. Computer-aided drug design (CADD) works sometimes and is improving quickly, partially thanks to deep learning. - Biological
Unlike chemically-derived small molecules, you don't create these, you find them in organisms in the soil, the ocean, etc. These molecules are generally more complex structurally than regular small molecules, since they've had millions of years to evolve that complexity. Many drugs come from natural sources, especially antibiotics; the main issue is how to find them. We have already found much of the low-hanging fruit, like penicillin, many times over. - DNA-encoded
It's obvious that you can use DNA to encode and evolve proteins. Less obviously, you can encode small molecules too, by chemically fusing DNA barcodes to your chemicals. These libraries are interesting because just like proteins, you can evolve them, but you retain the flexible chemistry of small molecules. DiCE Molecules, which launched last year, is doing this.
- Chemical
- Proteins
- Recombinant
You can make almost any protein recombinantly in a microbe, then use that to replace a missing (extracellular) enzyme. Proteins are big, and get degraded quickly in the gut by proteases, so you usually have to inject them. (If ingesting proteins had drug-like effects, then eating would be more hazardous!) They are also usually too large to enter cells efficiently, further limiting their use as drugs. - Antibodies
Antibodies are proteins, but with a specific template for interfacing with the immune system. They are very large and cannot enter cells, so they only target cell-surface or extracellular targets. That works extremely well for the 10-20% of proteins that are accessible that way. Antibodies are our closest thing to a magic bullet, and are among the most successful drugs of all time. You can evolve antibodies in mice, use phage display, or identify them in humans. There are several varieties: antibody fragments, nanobodies, BiTEs, etc. - Peptides
Peptides are basically just short proteins. They are interesting potential drugs because they exhibit some of the properties of small molecules (small size, sometimes able to enter cells) and some of the properties of proteins (safe, easy to synthesize). Peptides are still rapidly degraded in the gut, so most are injected. Some of the limitations of peptides being proteins can be overcome by using peptidomimetics like D-peptides. - Peptides (DNA-encoded)
Peptides are generally encoded by DNA anyway, but there is a nice way to evolve a library of peptides, analogous to DNA-encoded small molecules: the decades-old-but-still-futuristic phage display. You can use phage display to select for antibody fragment binding too. - Protein + nucleotide
CRISPR/Cas9 is a gene editing technology combining an enzyme that cuts DNA (Cas9) and an guide RNA that defines the target. Cas9 is a sophisticated enzyme, so it is quite large and cannot enter cells unaided. That means to make Cas9 into a real therapy you need a way to deliver it. Older gene editing technologies like TALENs and Zinc Fingers also work well if they can be delivered, though they are much harder to program than Cas9. - Protein + small molecule
Antibody-drug conjugates promise to combine the advantages of antibodies (targeting cells with high specificity) with small molecules (ability to enter cells). The canonical example is an antibody designed to find a cancer cell, then deliver an attached "warhead" small molecule to kill it. Stemcentrx is one of many companies working on ADCs.
- Recombinant
- Nucleotides
- mRNA
Instead of delivering proteins to cells, why not just deliver the instructions to make the protein? Unfortunately, just like proteins get degraded by proteases, RNA gets degraded by nucleases, so again it comes down to delivery. Like proteins, the body tolerates nucleotides very well. Moderna is developing several mRNA-based therapies. - Antisense RNA
Antisense RNA binds to mRNA and interferes with translation. There have been two successful antisense RNA therapies recently: one for Crohn's (mongersen, which can be orally administered, since Crohn's is a disease of the gut), and one for SMA (nusinersen, which is injected into the CSF). Despite not being the coolest technology around, antisense RNA has had some amazing successes. - Aptamers
Like antibodies, aptamers are evolved to bind their targets with high specificity, but instead of amino acids, they are made from DNA or RNA. Aptamers can be evolved using a simple process called SELEX (unfortunately IP-encumbered). As nucleotides, aptamers are degraded quickly and require special delivery mechanisms. The only approved aptamer therapy, Macugen, is for AMD and it's injected directly into the eye. - RNAi
RNAi encompasses a few related systems that are difficult for me to differentiate: siRNA, shRNA, miRNA, piRNA. RNA interference is an extremely effective tool for modifying cell lines, but so far has been less effective as a therapy. Alnylam has several RNAi therapies in development. - Raw DNA/RNA
Raw DNA or RNA with the right sequence will do gene editing at low frequency, even without a proper vector. You can also inject complete plasmids that get transcribed and translated (assuming some accompanying stress like electroporation). This is called a DNA vaccine, which is conceptually quite similar to an mRNA, except that DNA vaccines are injected with an adjuvant like Freund's adjuvant, to elicit an immune response to the resultant protein.
- mRNA
- Cells
Cell-based therapies are becoming increasingly important, especially for cancer treatment. In cancer, the idea is to train the immune system (usually T-cells) to attack cancer cells more efficiently. Since the vast majority of cancers are quashed by the immune system anyway, this makes a lot of sense. CAR-Ts — perhaps the best-known cell therapy for cancer — are T-cells that have been extracted and forced to express a cell-surface antibody fragment with specificity for a cancer, which forces the T-cell to engage the cancer. There's a lot more going on in cell-based therapy, like stem cell therapies, but I don't know too much about this.
Delivery
The big advantage of small molecules over proteins and nucleotides — and a watchword for most of the novel therapies above — is delivery: small molecules can get across the gut and penetrate the cell membrane to bind with targets inside the cell. Proteins and nucleotides are degraded quickly, especially in the gut, and proteins are usually way too large to enter cells anyway.
There are exceptions to this rule, cases when cells are more accessible: for example, the eye is easier to access than most organs (e.g., the aptamer therapy, Macugen); if your target is in the gut then you may be able to deliver it orally (e.g., the antisense RNA therapy, mongersen); if you can apply your therapy ex vivo (especially retransplantable tissue like liver or bone marrow) then you no longer need a delivery vector (e.g., Provenge and other adoptive cell transfers, gene editing in embryos). Examining the pipelines of the new batch of gene editing and RNA therapy biotechs, we see that the diseases they tackle are very often in the bone marrow, liver or eye.
If your target is inside a cell, the cell cannot be extracted for treatment ex vivo, and you can't develop a good small molecule, then what can you do? Many of the modern protein or nucleotide-based therapeutics listed above need some some additional help to protect them from degradation, localize them to target cells, and penetrate the cell membrane. It could be argued that delivery technology has lagged behind drug technology generally. There are still no great options for delivery.
- Viruses
Viruses, especially AAV, are especially useful for nucleotide therapies like gene therapy. This makes sense when you consider that viruses are nanomachines, evolved over billions of years to target cells and inject nucleotides. However, since viruses are often immunogenic, there can be side-effects. - Nanoparticles / Liposomes
In rare cases, liposomes have been used to deliver drugs, like cisplatin. We were recently surprised to learn that Moderna, the mRNA biotech, is using lipid-nanoparticles to deliver mRNA to cells. Virus-like particles (essentially noninfectious viruses) may combine the best of both worlds.
For a more detailed discussion on delivery methods, there's a good recent review in NRDD.