Using GPT-3 as a knowledge-base in biotech
By now most people have seen what Chat-GPT / GPT-3 can do.
It's an amazing tool, but it has a loose affiliation with the truth.
This makes it great for anything creative like generating ideas or filler text,
but less useful if you are trying to search for information.

Tools like GPT Index (now LlamaIndex) allow you to add additional text to augment GPT-3. This leads to the obvious idea: what if you could create a custom knowledge-base for your lab or biotech, indexing all scientific papers, assay results, and written communications. This could be an extremely powerful tool for scientists to access insitutional knowledge, find buried information, etc.
You can get GPT Index running very quickly by following the quickstart. However, for me, trained on scientific papers, it performed worse than vanilla Chat-GPT, even for facts directly addressed in the corpus.
Paper QA
GPT Index is a powerful, but general tool. For our purposes, Andrew White's Paper QA improves upon it by adding the important concept of citations. Citations turn out to be extremely useful for keeping the model honest, and identifying the source of the information for follow-up or verification.
There are two main ways of using this tool:
- Automatically search for papers relevant to your question (e.g., on Google Scholar), index those papers, and respond (as Andrew White does)
- Index a large corpus of all papers relevant to your lab or biotech once, and respond referencing that corpus
Here, I attempt to get option 2 working. I took a set of 200 scientific papers, indexed them, and tested the answers compared to Chat-GPT.
Results
It's difficult to quantify the results here, but compared to Chat-GPT, Paper QA was less verbose (sometimes good, sometimes bad), less prone to invention, and had explicit citations. Sometimes Paper QA even usefully responds with "I cannot answer".
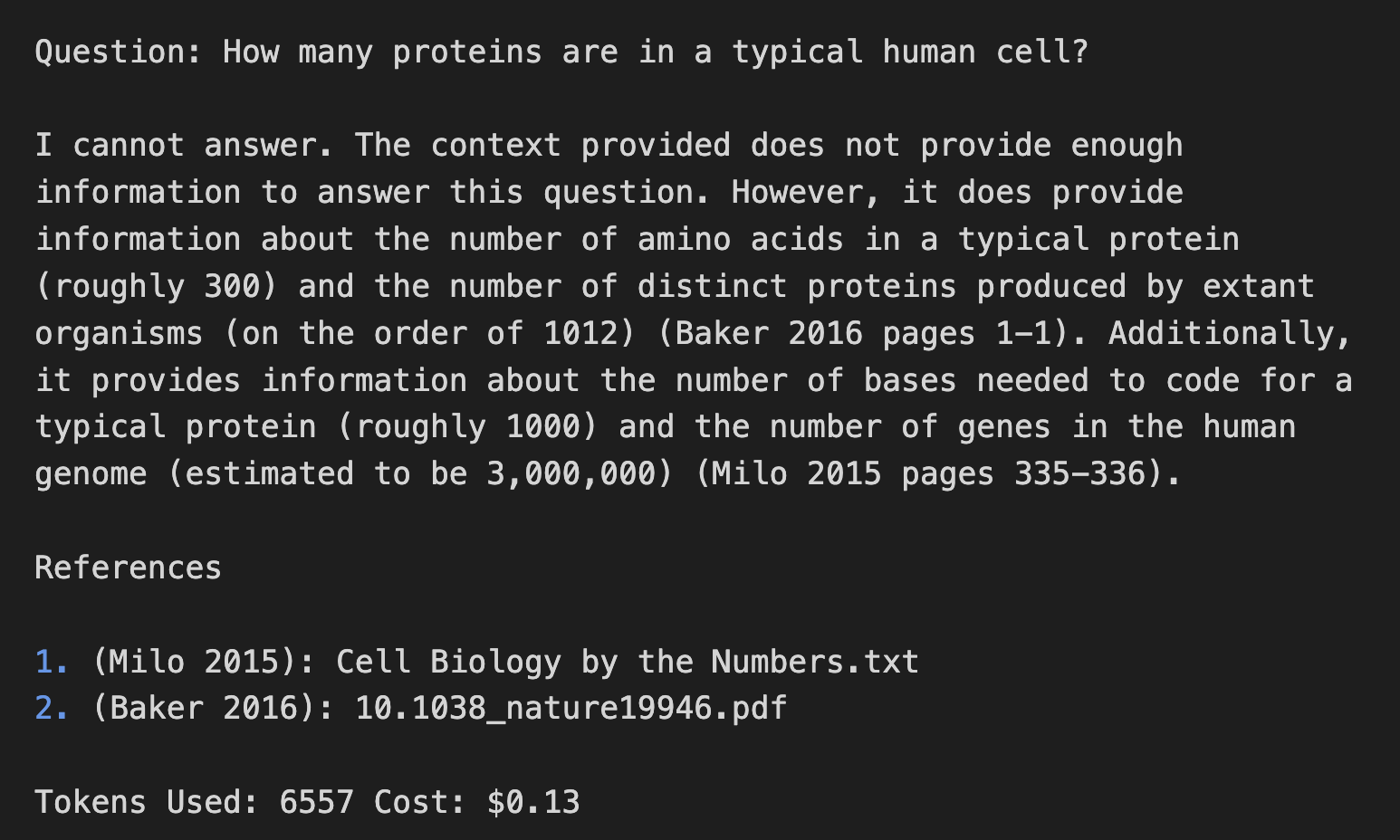
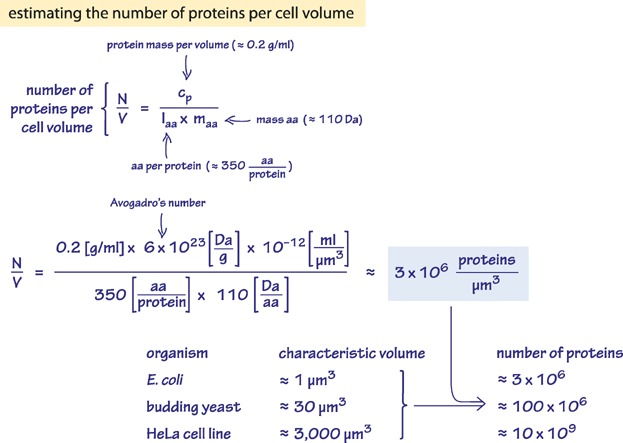
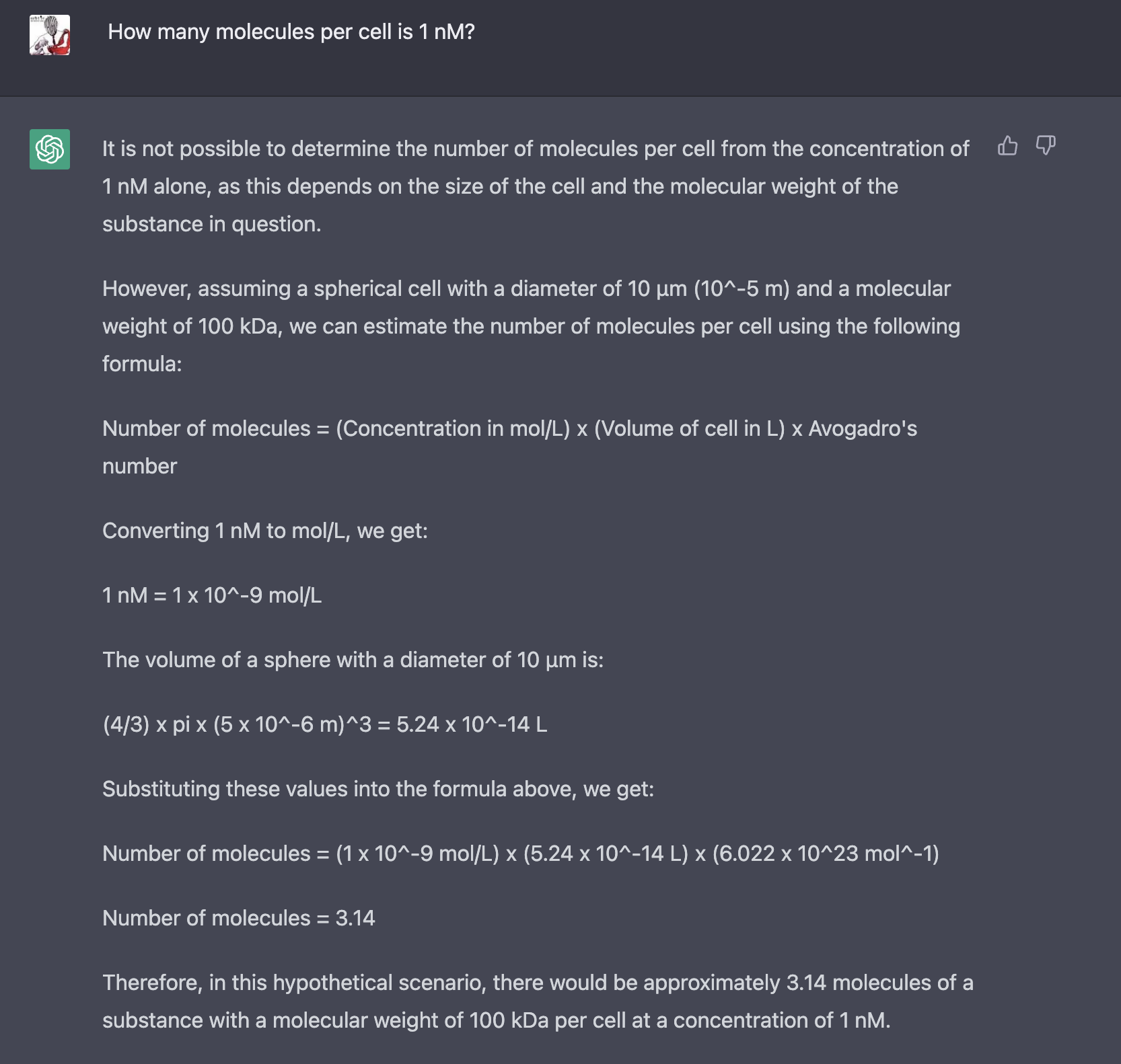
Trained on Cell Biology By The Numbers, it does a pretty good job of answering, and giving the relevant page numbers. However, the answers are far from perfect, and include some incorrect statements like "3 million genes in the human genome". (Maybe it is dividing the size of the human genome by the size of the "typical protein"?) Chat-GPT gives more detailed answers, but some of the details are strange, and though the formulas are right, the calculations are off by an order of magnitude (twice!).


Issues
All of this stuff is very new, so there are limitations.
-
The system as implemented costs around 15c per query. Not terrible, but if you had hundreds of users you'd have to pay attention to costs.
-
Even with the addition of citations, it is still prone to fabulation, just like Chat-GPT.
-
Chat-GPT can often produce results that feel more complete, depending on the type of question. Additionally, you can correspond with Chat-GPT, asking follow-on questions or clarifications.
-
You cannot index e.g., your Slack history or your proprietary databases yet, since OpenAI will retain access to all uploaded text. Once an open-source language model emerges (actually open, not pretend open like LlaMa), the addition of this kind of data could make the search really interesting though — we could even ask questions about results stored in the database!
Conclusions
The system works well on certain types of tasks:
- If you read something a while back, and want to remind yourself what it was and where you read it
- If you want a very short summary / answer on some topic referenced in the corpus, and you are willing to check the reference to validate
Overall, this seems like a great tool for searching a library of PDFs, though anecdotally, the performance may degrade somewhat as you add papers (e.g., to get a summary of one paper, it may be better to index only that paper.) I can see using the tool routinely, but the next stage, where we index everything anyone has typed, read, or generated in an organization could be where things get very exciting. Or maybe we just go full multimodal and learn a language model for all of life!
Code
You can easily create your own scientific paper knowledge-base with the following code.
For simplicity, I make the citation derive from the filename of the PDF,
with all PDFs ending in _[Authorname]_[Year].pdf.
(Note Authorname must be capitalized for Paper QA!)
You could also use Zotero's bibliography output or similar.
index_pdfs.py
'''
Script to index a directory of PDFs for GPT queries
You need an OpenAI API key to run it.
Register and get a key at https://platform.openai.com/account/api-keys
Uses https://github.com/whitead/paper-qa
Before running the script, install paper-qa:
```pip install paper-qa```
Then run:
```python index_pdfs.py $PDF_DIR```
The cost to build the index should be around ~1c per paper. It cost me <$2 to index 200 papers.
The code indexes 30 papers at a time by default, since otherwise you may hit usage limits.
You can add to the index by just running `python index_pdfs.py` multiple times,
or just changing the limit.
Please check https://platform.openai.com/account/usage to monitor costs!
'''
from paperqa import Docs
import glob, os, pickle, sys
os.environ["OPENAI_API_KEY"] = ADD_OPENAI_API_KEY_HERE
PKL = "pdf_index.pkl"
MAX_PAPERS_PER_RUN = 30
pdfs = glob.glob(f"{sys.argv[1]}/*.pdf")
if os.path.exists(PKL) and os.path.getsize(PKL) > 0:
with open(PKL, "rb") as f:
docs = pickle.load(f)
else:
docs = Docs()
papers_added = 0
for pdf in pdfs:
citation = ' '.join(pdf.split('.')[-2].split('_')[-2:])
if os.path.basename(pdf) in [os.path.basename(f) for f in docs.docs.keys()]:
continue
print(f"Indexing {citation} {pdf}")
try:
docs.add(pdf, citation)
papers_added += 1
if papers_added >= MAX_PAPERS_PER_RUN:
break
except Exception as e:
print("ERR", e, pdf)
with open(PKL, "wb") as f:
pickle.dump(docs, f)
query_pdfs.py
'''
Script to ask questions of your indexed PDFs using GPT.
Run, e.g.:
```python query_pdfs.py What is the largest mammal?```
'''
from paperqa import Docs
import os, pickle, sys
os.environ["OPENAI_API_KEY"] = ADD_OPENAI_API_KEY_HERE
PKL = "pdf_index.pkl"
with open(PKL, "rb") as f:
docs = pickle.load(f)
question = ' '.join(sys.argv[1:])
answer = docs.query(question)
print(answer)