I really like resampling methods for data analysis, and I think they are under-utilized. The main thing these techniques have in common is that instead of an analytical solution, they make a computer repeatedly analyze variations of the data. Examples of resampling methods include permutation tests, the bootstrap, and cross-validation.
Cross-validation in particular seems to have really taken over model selection over the last ten years or so (e.g., compared to the more "statistical" AIC or BIC). There may be an argument here that this is due to non-statisticians needing statistical tests (e.g., in machine learning) so they naturally gravitate to using computational power instead of statistical expertise.
In biology, there are a few tests than come up a lot, but the main one is probably testing if distribution A is different to distribution B (e.g., experiment vs control). This is usually done with a t-test, but can also be done — with some advantages — by a permutation test. I looked for a post on this topic but could not find one that exactly covered what I was interested in, so this article will show some data on this.
Here are a few related articles that may be of interest:
- The Permutation Test: some nice animations showing the basics of permutation tests
- tidy resampling: using a permutation test to replace a t-test, in tidy R
- Why Permutation Tests are Superior to t and F Tests in Biomedical Research: "Because randomization rather than random sampling is the norm in biomedical research and because group sizes are usually small, exact permutation or randomization tests for differences in location should be preferred to t or F tests."
- Common statistical tests are linear models: only related in theme, but has t-tests in there, and worth reading
The t-test
I would guess most biologists use t-tests occasionally, as the simplest way to check if an experimental result differs from a control. I would also argue that most people who use the t-test don't really know the main assumptions:
- the distributions are normal
- the variances of the distributions are equal
- the sample size is large
If you violate any of these assumptions, you are supposed to consider another test.
If distributions are not normal
We usually assume if a distribution is not normal then it's some unknown distribution. In this case you could use a non-parametric test, like the Mann–Whitney U test. Of course, there's no such thing as a completely normal distribution, so it's a bit unclear where to draw the line here.
If variances are unequal
If the variances are unequal, Welch's t-test is recommended. Welch's t-test is "an adaptation of Student's t-test, and is more reliable when the two samples have unequal variances and/or unequal sample sizes." I'm not sure I've ever seen this test used in a biology paper, though variances are very often unequal. For example, any time you are counting things (colonies, etc) it's likely a Poisson distribution where the variance equals the mean.
If sample size is small
It's a bit of a grey area how few samples count as "small", but some sources say below 30. That is actually a pretty large number for many biological experiments!
For example, if I do a quick review of the first four papers that pop up for the search "xenograft tumor treatment t-test":
- https://acsjournals.onlinelibrary.wiley.com/doi/full/10.1002/cncr.22441: <=20 mice
- https://cancerres.aacrjournals.org/content/61/20/7669.full-text.pdf: "Data (n = 3) were compared with VEGF controls using t test analysis"
- https://cancerres.aacrjournals.org/content/67/8/3560.full-text.pdf: 7 groups of 7 mice
- https://link.springer.com/article/10.1007/s10585-010-9321-4: 10 mice
In order to publish, the authors of these papers need to be able to report a p-value, and the t-test is the most convenient, and accepted tool.
Violations
It's possible that the above violations in assumptions are just fine, and the stats work out fine. Still, the lack of clarity on what counts as a violation here seems to defeat the purpose of having the rules, and a permutation test might offer a safer approach.
Permutation Test
The general problem below will be to determine if two distributions with mean of 0 (red) and diff (yellow), are different.
from scipy.stats import norm, laplace, ttest_ind, median_test, mannwhitneyu, pearsonr, linregress
from matplotlib import pyplot as plt
from IPython.display import SVG
import seaborn as sns
import pandas as pd
import numpy as np
from tqdm.auto import tqdm
tqdm.pandas()
f, ax = plt.subplots(figsize=(12,4))
diff = 1
dist1 = norm(0, 1).rvs(100)
dist2 = norm(diff, 1).rvs(100)
sns.histplot(dist1, color='r', bins=40);
sns.histplot(dist2, color='y', bins=40);
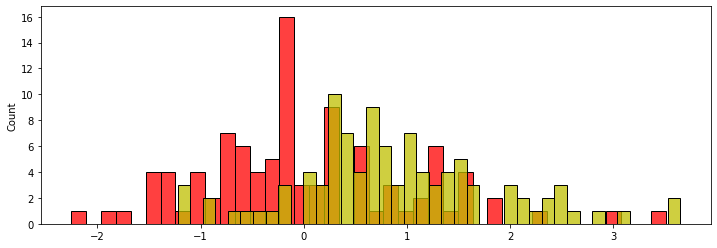
This is the code for a permutation test, for use in place of a one- or two-sided t-test, Welch's t-test, etc. In all examples below I will use a two-sided t-test, since this is more commonly used than one-sided. (Although to me, one-sided makes more sense most the time!)
def ptest_ind(dist1, dist2, fn=np.mean, N=30000, alternative="two-sided"):
"""permutation test, matching scipy.ttest_ind syntax"""
from collections import namedtuple
from warnings import warn
PTResult = namedtuple("PTResult", ["stat", "pvalue"])
new_dists = np.concatenate([dist1, dist2])
test = fn(dist1) - fn(dist2)
res = []
for _ in range(N):
np.random.shuffle(new_dists)
ndist1 = new_dists[:len(dist1)]
ndist2 = new_dists[len(dist1):]
res.append(test > fn(ndist1) - fn(ndist2))
stat = sum(res) / N
if alternative == "less":
pvalue = stat
elif alternative == "greater":
pvalue = 1 - stat
elif alternative == "two-sided":
pvalue = 2*min(stat, 1-stat)
else:
raise ValueError(f"{alternative=}")
if pvalue == 0:
warn(f"permutation test sample limit reached: <1/{N}")
pvalue = 1.0/N
return PTResult(stat, pvalue)
def plot_corr(df, x, y, min_xy):
"""Plot the correlation between two tests, and report Pearson's r"""
lx, ly = f"log_{x}", f"log_{y}"
_df = (df_rows
.replace([np.inf, -np.inf], np.nan)
.dropna()
.assign(**{lx: lambda df: df[x].apply(np.log10), ly: lambda df: df[y].apply(np.log10)}))
res = linregress(_df[ly], _df[lx])
f, ax = plt.subplots(figsize=(6,6))
sns.scatterplot(data=_df, x=lx, y=ly, ax=ax);
sns.lineplot(x=np.array([0, min_xy]), y=res.intercept + res.slope*np.array([0, min_xy]), color='r', linewidth=0.5, ax=ax)
ax.set_xlim(min_xy, 0);
ax.set_ylim(min_xy, 0);
ax.axline((min_xy, min_xy), (0, 0), linewidth=0.5, color='g');
return res.rvalue
t-test with normal distributions
This is the simplest case: comparing two equal variance, high sample size, normal distributions. Here we see an almost perfect correspondence between the results of the t-test and permutation test. This is good, since it shows the permutation test is not reporting biased (e.g., too low) p-values.
sample_size = 30
rows = []
for diff in tqdm(np.tile(np.arange(0, 0.4, 0.02), 1)):
dist1 = norm(0, 1).rvs(sample_size)
dist2 = norm(diff, 1).rvs(sample_size)
tt = ttest_ind(dist1, dist2, alternative="two-sided").pvalue
pt = ptest_ind(dist1, dist2, alternative="two-sided").pvalue
rows.append((tt, pt))
df_rows = pd.DataFrame(rows, columns=["tt", "pt"])
print(f't-test vs permutation r: {plot_corr(df_rows, "tt", "pt", -3):.3f}')
t-test vs permutation r: 1.000

t-test with normal distributions, but low sample size
What happens to the results of a t-test when the number of samples is low? Here, the low sample size results in some noise that decreases the correlation, but it still seems to work very similarly to the permutation test, even with just 3 samples (despite the regression line below).
sample_size = 3
rows = []
for diff in tqdm(np.tile(np.arange(0, 0.6, 0.03), 5)):
dist1 = norm(0, 1).rvs(sample_size)
dist2 = norm(diff, 1).rvs(sample_size)
tt = ttest_ind(dist1, dist2, alternative="two-sided").pvalue
pt = ptest_ind(dist1, dist2, alternative="two-sided").pvalue
rows.append((tt, pt))
df_rows = pd.DataFrame(rows, columns=["tt", "pt"])
print(f't-test vs permutation r: {plot_corr(df_rows, "tt", "pt", -2):.3f}')
/tmp/ipykernel_2790/1233266942.py:30: UserWarning: permutation test sample limit reached: <1/30000
warn(f"permutation test sample limit reached: <1/{N}")
t-test vs permutation r: 0.760

t-test with normal distributions, but unequal variances
scipy's ttest_ind has an option to use Welch's t-test instead of a t-test.
Again, the results are almost identical between Welch's t-test and the permutation test.
sample_size = 30
rows = []
for diff in tqdm(np.tile(np.arange(0, 1.5, 0.075), 5)):
dist1 = norm(0, 1).rvs(sample_size)
dist2 = norm(diff, 2).rvs(sample_size)
tt = ttest_ind(dist1, dist2, alternative='two-sided', equal_var=False).pvalue
pt = ptest_ind(dist1, dist2, alternative="two-sided").pvalue
rows.append((tt, pt))
df_rows = pd.DataFrame(rows, columns=["tt", "pt"])
print(f't-test vs permutation r: {plot_corr(df_rows, "tt", "pt", -3):.3f}')
/tmp/ipykernel_2790/1233266942.py:30: UserWarning: permutation test sample limit reached: <1/30000
warn(f"permutation test sample limit reached: <1/{N}")
t-test vs permutation r: 0.967

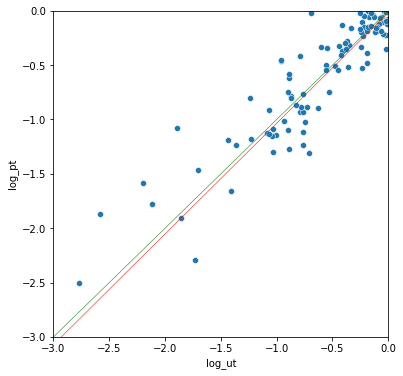
t-test with non-normal distributions
In this case, the Mann-Whitney U test is usually recommended instead of the t-test. The Mann-Whitney U test "is a nonparametric test of the null hypothesis that, for randomly selected values X and Y from two populations, the probability of X being greater than Y is equal to the probability of Y being greater than X." However, two distributions can have equal medians, but different distributions, and Mann-Whitney may return a significant p-value.
Another option is to directly test for a difference in medians, with Mood's median test.
I'm not sure why the permutation test and median test results don't match more closely here, though the median test only produces a few possible p-values. Still, the results are quite close, and appear unbiased. The correlation between the permutation tests and Mann-Whitney U test is also very high.
sample_size = 30
rows = []
for diff in tqdm(np.tile(np.arange(0, 0.5, 0.025), 5)):
dist1 = laplace(0, 1).rvs(sample_size)
dist2 = laplace(diff, 1).rvs(sample_size)
mt = median_test(dist1, dist2, ties="ignore")[1] # ugh, inconsistent scipy syntax
ut = mannwhitneyu(dist1, dist2, alternative="two-sided")[1] # ugh, inconsistent scipy syntax
pt = ptest_ind(dist1, dist2, fn=np.median, alternative="two-sided").pvalue
rows.append((mt, ut, pt))
df_rows = pd.DataFrame(rows, columns=["mt", "ut", "pt"])
print(f'median test vs permutation r: {plot_corr(df_rows, "mt", "pt", -3):.3f}')
print(f'mann-whitney vs permutation r: {plot_corr(df_rows, "ut", "pt", -3):.3f}')
print(f'mann-whitney vs median test r: {plot_corr(df_rows, "ut", "mt", -3):.3f}')
median test vs permutation r: 0.863
mann-whitney vs permutation r: 0.920
mann-whitney vs median test r: 0.873


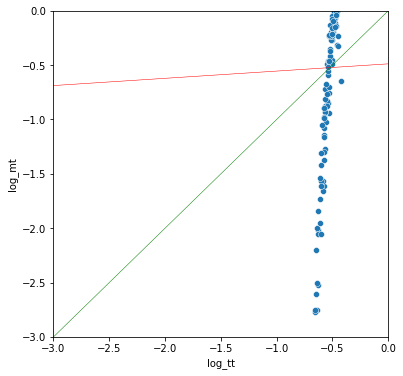
t-test with normal distributions, but one outlier sample
A common problem with real-world datasets is outlier datapoints that greatly skew the distribution and hence skew p-values. Removing or accounting for outliers will increase the robustness of a test (e.g., how sensitive the result is to an erroneous reading).
It's interesting that the t-test is reasonably well correlated with the other two tests, but way off the diagonal.
sample_size = 30
rows = []
for diff in tqdm(np.tile(np.arange(0, 0.5, 0.025), 5)):
dist1 = np.concatenate([norm(0, 1).rvs(sample_size), np.array([0])])
dist2 = np.concatenate([norm(diff, 1).rvs(sample_size), np.array([100])])
mt = mannwhitneyu(dist1, dist2, alternative='two-sided').pvalue
tt = ttest_ind(dist1, dist2, alternative="two-sided").pvalue
pt = ptest_ind(dist1, dist2, fn=np.median, alternative="two-sided").pvalue
rows.append((mt, tt, pt))
df_rows = pd.DataFrame(rows, columns=["mt", "tt", "pt"])
print(f'mann-whitney vs permutation r: {plot_corr(df_rows, "mt", "pt", -3):.3f}')
print(f't-test vs mann-whitney r: {plot_corr(df_rows, "tt", "mt", -3):.3f}')
print(f't-test vs permutation r: {plot_corr(df_rows, "tt", "pt", -3):.3f}')
mann-whitney vs permutation r: 0.923
t-test vs mann-whitney r: 0.926
t-test vs permutation r: 0.835


Conclusion
In general, the permutation test gives you the same answer you would get if you chose the "right" test for your task. But, you have to exert less effort in figuring out what the right test is.
There is a very direct connection between the test you want and the calculation performed. For example, with the Mann-Whitney U test, it's confusing what is actually being tested, apart from "the non-parametric alternative to a t-test".
The permutation test is also robust in a way that serves most use-cases. Outliers are handled conservatively, and there are few or no assumptions to violate.
Disadvantages
There are some disadvantages too, but relatively minor I think.
- Regular t-tests do pretty well in my tests above, even with only 3 samples! The only major problem is with outliers, which can imply extremely wide distributions if taken at face-value.
- Permutation tests can be slow, so if you are doing thousands of tests, or you need precise, very low p-values, then they will likely not be suitable.
- You have to know precisely what you are testing. For example, "are the medians of these two distributions identical?". This is mostly an advantage in that it's more explicit.
- You get more statistical power if you use the test that includes the correct assumptions. This seems to be a very minor effect in my tests above.
Overall, you can't go too far wrong with permutation tests since they are so simple. If nothing else, they are a good way to sanity check results from other tests with more assumptions.