In a previous blogpost I described a pipeline for synthesizing arbitrary proteins on the transcriptic robotic lab platform using only Python code. The ultimate goal of that project was to be able to run a program that takes a protein sequence as input, and "returns" a tube of bacteria expressing that protein. Here I'll describe some progress towards that goal.
Pipelining
The usual way to chain together different programs in bioinformatics is with a pipeline management system, for example, snakemake, nextflow, toil, WDL, and many many more. I've recently become a big fan of nextflow for computational pipelines, but its major advantages (e.g., containerization) don't help much here because so much of the work happens outside of the computer. For this project I've been using the slightly simpler snakemake, mainly for tracking which steps have been completed, and deciding which steps can be run in parallel based on their dependencies.
Each protocol has four associated steps in the pipeline:
- generate protocol: create an autoprotocol file describing the protocol
- submit protocol: submit the autoprotocol file to transcriptic
- get results: download images, data, etc. from transcriptic
- create report: create a HTML report from the downloaded data
snakemake pipeline for protein synthesis
Metaprotocol
In my terminology, a "metaprotocol" defines the complete process, which is turned into a series of protocols. Ideally, the output of a single protocol will be a decision point: for example, whether or not a gel image includes the expected bands.
The metaprotocol is defined in yaml, which has its issues, but is more readable than json, and well supported. This code depends heavily on Pydna, a Python package for cloning and assembly. Given an insert and a vector, Pydna will design primers and a PCR program. The following is my metaprotocol yaml for expressing GFP:
- meta: assembly: |- Assembly: Sequences........................: [2690] [786] Sequences with shared homologies.: [2690] [786] Homology limit (bp)..............: 25 Number of overlaps...............: 2 Nodes in graph(incl. 5' & 3')....: 4 Only terminal overlaps...........: No Circular products................: [3412] Linear products..................: [3446] [3442] [34] [30] assembly_figure: |2- -|SYNPUC19V|31 | \/ | /\ | 31|786bp_PCR_prod|30 | \/ | /\ | 30- | | --------------------------------- metaprotocol_id: 1k9ginus pcr_figure: |2- 5AGGAGGACAGCTATGTCGAAAGGA...CATTACCCATGGAATGGATGAACTGTATAAA3 ||||||||||||||||||||||||||||||| tm 59.8 (dbd) 70.6 3GTAATGGGTACCTTACCTACTTGACATATTTTTAAGTGACCGGCAGCAAAATGTTGCAGCA5 5ACTCTAGAGGATCCCCGGGTACCGAGCTCGAGGAGGACAGCTATGTCGAAAGGA3 |||||||||||||||||||||||| tm 62.1 (dbd) 69.3 3TCCTCCTGTCGATACAGCTTTCCT...GTAATGGGTACCTTACCTACTTGACATATTT5 pcr_program: |2 Pfu-Sso7d (rate 15s/kb) Two-step| 30 cycles | |786bp 98.0°C |98.0C | |Tm formula: Pydna tmbresluc _____ __|_____ | |SaltC 50mM 00min30s|10s \ | |Primer1C 1.0µM | \ 72.0°C|72.0°C|Primer2C 1.0µM | \______|______|GC 49% | 0min11s|10min |4-12°C project_name: pUC19_sfGFP_cloning_v1 - linearize: restriction_enzyme: EcoRI vector: pUC19 - oligosynthesize: p1: ACTCTAGAGGATCCCCGGGTACCGAGCTCGAGGAGGACAGCTATGTCGAAAGGA p2: ACGACGTTGTAAAACGACGGCCAGTGAATTTTTATACAGTTCATCCATTCCATGGGTAATG - thermocycle: insert: AGGAGGACAGCTATGTCGAAAGGAGAAGAACTGTTTACCGGTGTGGTTCCGATTCTGGTAGAACTGGATGGGGACGTGAACGGCCATAAATTTAGCGTCCGTGGTGAGGGTGAAGGGGATGCCACAAATGGCAAACTTACCCTTAAATTCATTTGCACTACCGGCAAGCTGCCGGTCCCTTGGCCGACCTTGGTCACCACACTGACGTACGGGGTTCAGTGTTTTTCGCGTTATCCAGATCACATGAAACGCCATGACTTCTTCAAAAGCGCCATGCCCGAGGGCTATGTGCAGGAACGTACGATTAGCTTTAAAGATGACGGGACCTACAAAACCCGGGCAGAAGTGAAATTCGAGGGTGATACCCTGGTTAATCGCATTGAACTGAAGGGTATTGATTTCAAGGAAGATGGTAACATTCTCGGTCACAAATTAGAATACAACTTTAACAGTCATAACGTTTATATCACCGCCGACAAACAGAAAAACGGTATCAAGGCGAATTTCAAAATCCGGCACAACGTGGAGGACGGGAGTGTACAACTGGCCGACCATTACCAGCAGAACACACCGATCGGCGACGGCCCGGTGCTGCTCCCGGATAATCACTATTTAAGCACCCAGTCAGTGCTGAGCAAAGATCCGAACGAAAAACGTGACCATATGGTGCTGCTGGAGTTCGTGACCGCCGCGGGCATTACCCATGGAATGGATGAACTGTATAAA p1: ACTCTAGAGGATCCCCGGGTACCGAGCTCGAGGAGGACAGCTATGTCGAAAGGA p2: ACGACGTTGTAAAACGACGGCCAGTGAATTTTTATACAGTTCATCCATTCCATGGGTAATG program: extension_time: 11.0 forward_primer_concentration: 0.001 rate: 15.0 reverse_primer_concentration: 0.001 saltc: 50.0 ta: 72.0 - assemble: insert: sfGFP vector: pUC19
DNA synthesis
Of course, before you can run this pipeline, you need to have the appropriate insert DNA in your transcriptic inventory. As far as I know, none of the major synthetic DNA suppliers has an API. However, you can order DNA from IDT by filling in an excel file. I have automated filling in and emailing this file, so DNA synthesis can be included in the pipeline too! It should take about a week from ordering for DNA to appear at transcriptic.
Reporting
After each protocol finishes, a HTML report is generated. This allows the user to evaluate protocol results manually before initiating the next step. There are ways to automate this more, like using automated band mapping of gel images, but I think that kind of thing will work better once the transcriptic API settles down a bit. The HTML report also serves as a log of the experiment.
cut_plasmid FINISHED
Submitted at UTC 2016-08-20 19:42:48 Started at UTC 2016-08-20 22:52:06 Completed at UTC 2016-08-21 01:28:49 Ran report at UTC 2016-10-26 22:15:03
Expected DNA bands of size: 2686bp
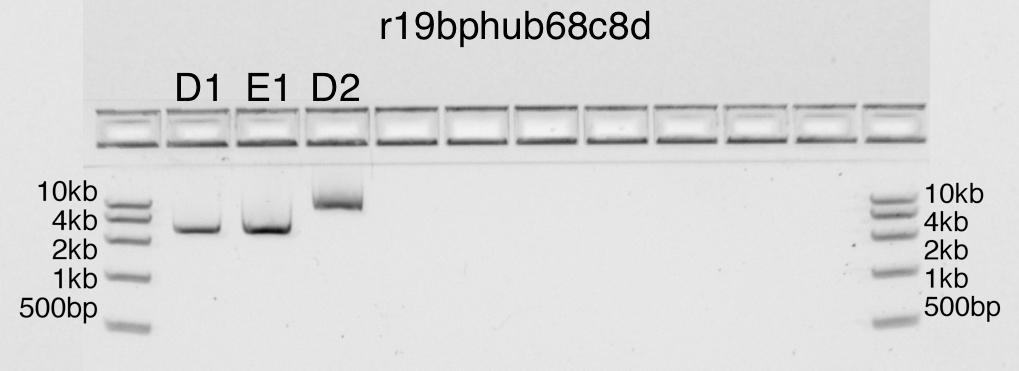
- runstatus:
- cut_plasmid_pUC19_EcoRI.runstatus.json
- sentinel:
- cut_plasmid_pUC19_EcoRI.sentinel
synthesize_primers FINISHED
Submitted at UTC 2016-08-23 19:35:37 Started at UTC 2016-08-23 20:00:11 Completed at UTC 2016-08-24 20:01:03 Ran report at UTC 2016-10-26 22:15:44
Synthesized primers:
ACTCTAGAGGATCCCCGGGTACCGAGCTCGAGGAGGACAGCTATGTCGAAAGGA ACGACGTTGTAAAACGACGGCCAGTGAATTTTTATACAGTTCATCCATTCCATGGGTAATG
- sentinel:
- synthesize_primers_pUC19_sfGFP.sentinel
add_flanks FINISHED
Submitted at UTC 2016-10-08 15:12:38 Started at UTC 2016-10-10 22:22:16 Completed at UTC 2016-10-11 01:04:12 Ran report at UTC 2016-11-17 15:36:02
PCR program
Pfu-Sso7d (rate 15s/kb)
Two-step| 30 cycles | |786bp
98.0°C |98.0C | |Tm formula: Pydna tmbresluc
_____ __|_____ | |SaltC 50mM
00min30s|10s \ | |Primer1C 1.0µM
| \ 72.0°C|72.0°C|Primer2C 1.0µM
| \______|______|GC 49%
| 0min11s|10min |4-12°C
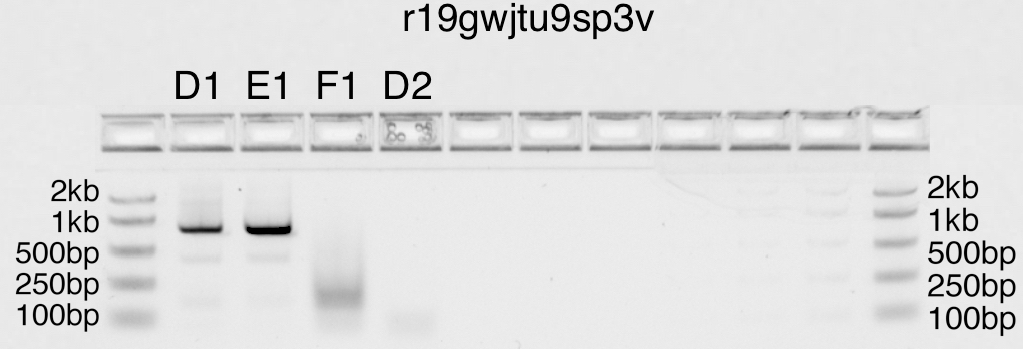
- runstatus:
- add_flanks_pUC19_sfGFP.runstatus.yaml
run_gibson_and_transform FINISHED
Submitted at UTC 2016-10-24 23:10:06 Started at UTC 2016-10-28 17:45:56 Completed at UTC 2016-10-29 17:01:26 Ran report at UTC 2016-10-30 15:12:18
pick_colonies_and_culture FINISHED
Submitted at UTC 2016-11-02 19:46:39 Started at UTC 2016-11-04 00:11:21 Completed at UTC 2016-11-04 17:54:20 Ran report at UTC 2016-11-11 01:10:22
Absorbance readings
pick_colonies_and_culture_v1_abs_t16 b1 0.047 a2 0.048 a10 0.050 a7 0.050 a6 0.051 a5 0.051 b6 0.054 b5 0.056 a11 0.058 a3 0.060 b7 0.066 a9 0.071 a1 0.073 b4 0.080 a12 0.080 b10 0.081 a4 0.082 b9 0.083 b2 0.086 b3 0.088 a8 0.090 b11 0.116 b8 0.148
- abs_data:
- pick_colonies_and_culture.abs_data.json
- runstatus:
- pick_colonies_and_culture.runstatus.yaml
Conclusions
There is still plenty to do before the pipeline is completely automatic. For example, attentive readers will notice that the HTML report above shows an unsuccessful transformation, one of many! The first complete transformation took several months to get right. The biggest challenge is making the process robust to changes in the protein sequence — even basic PCR can go wrong in many ways. Currently, debugging is a major undertaking; unlike regular programming, iterations are slow and expensive. However, if the protocols can be made robust enough, which I think they can, then synthesizing a new protein could become as simple as running BLAST.
